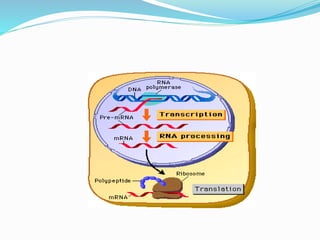
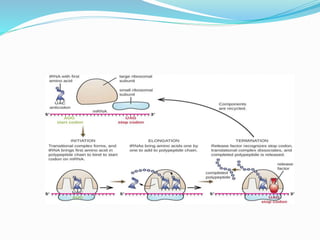
The document summarizes the process of translation. It describes:
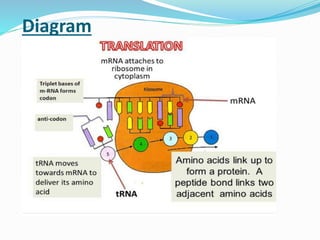
1) The machinery involved including mRNA, tRNA, ribosomes and other proteins.
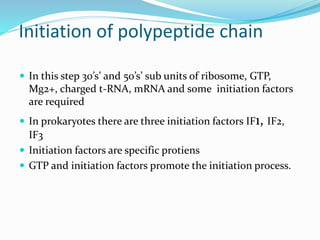
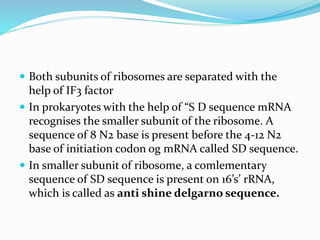
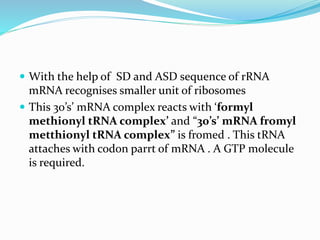
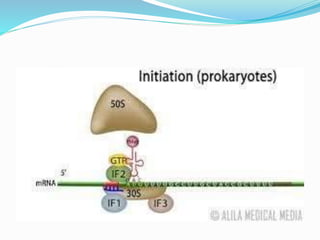
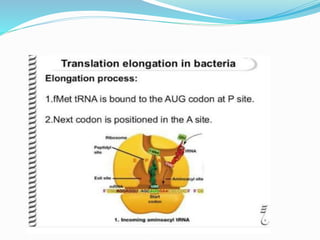
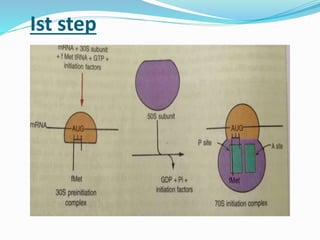
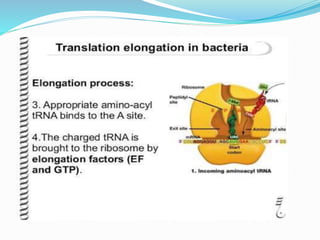
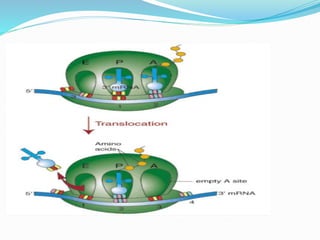
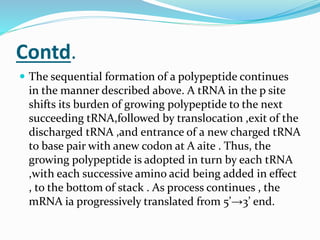
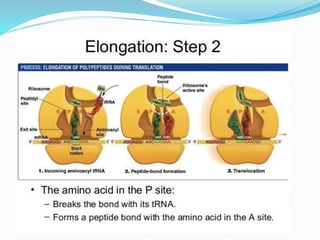
2) The three main steps - initiation, elongation, and termination. Initiation involves binding of the ribosome and first tRNA. Elongation is the repetitive addition of amino acids by tRNA and peptide bond formation. Termination occurs when a stop codon is reached and release factors trigger the release of the complete protein.
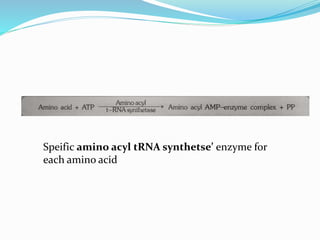
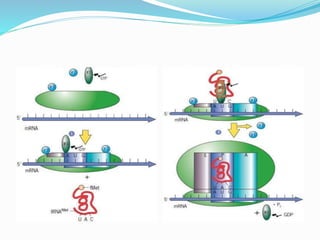
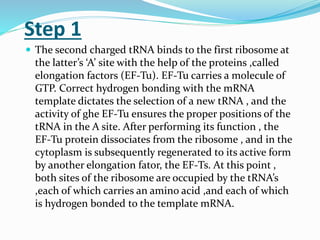
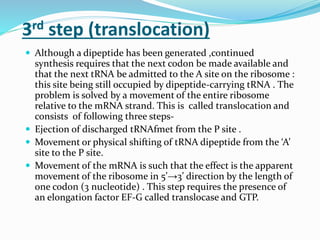
3) Key processes within each step like activation of amino acids, charging of tRNA, translocation during elongation, and hydrolysis of the peptide bond during termination.