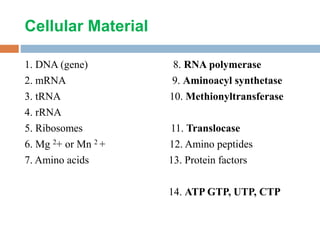
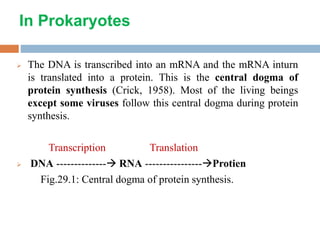
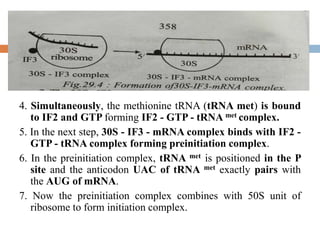
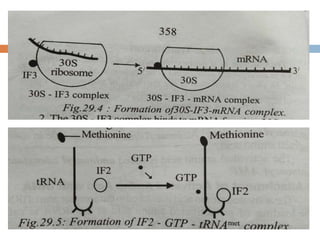
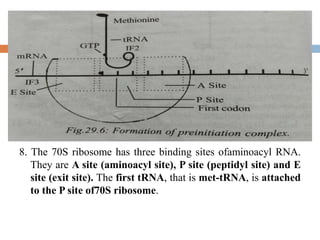
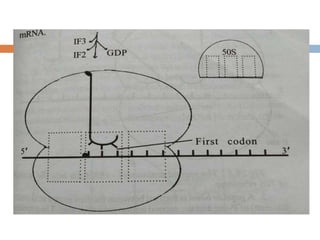
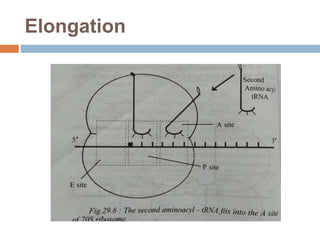
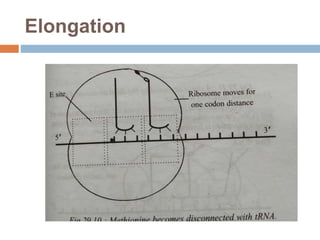
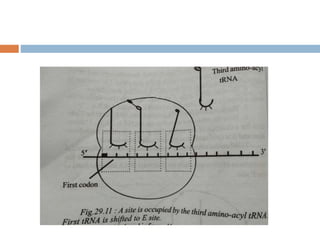
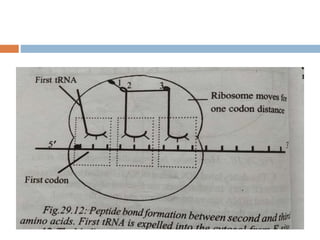
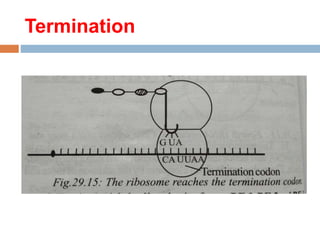
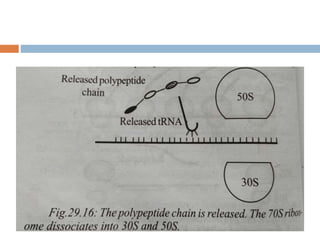
Protein synthesis is the process of constructing protein molecules from amino acids through transcription and translation, occurring primarily in the cytoplasm. The central dogma involves the transcription of DNA to mRNA, followed by the translation of mRNA into a polypeptide chain, requiring various cellular components including ribosomes and tRNA. The process also includes post-translational modifications to activate the polypeptide chain after synthesis.