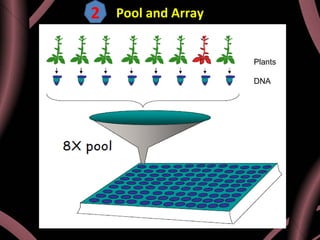
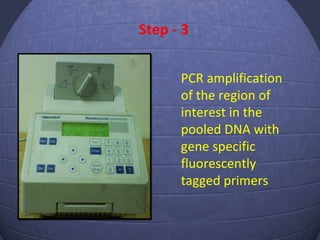
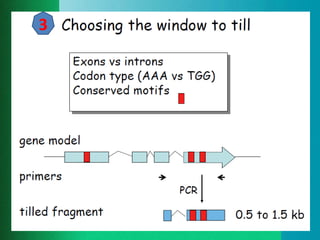
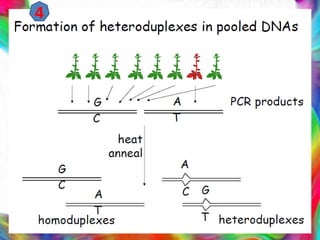
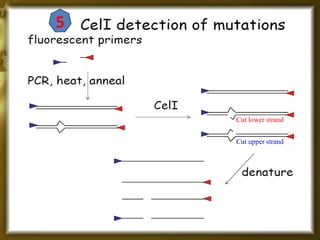
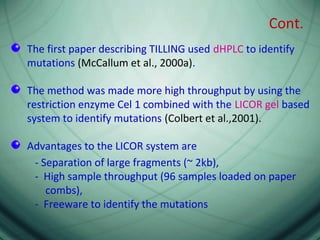
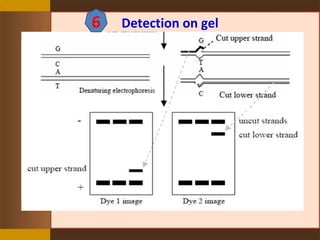
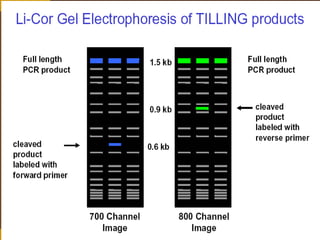
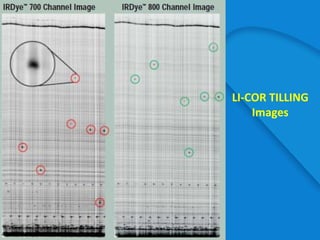
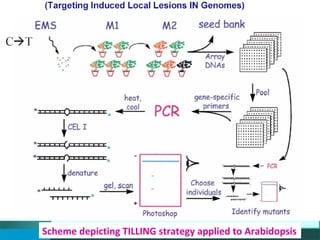
This document provides an outline for a presentation on Targeting Induced Local Lesions In Genomes (TILLING). TILLING is a technique that combines chemical mutagenesis with high-throughput screening to induce and identify point mutations in genes of interest. The presentation covers the principle, steps, applications, merits, and demerits of the TILLING technique. It involves chemically mutagenizing an organism using EMS, screening pooled DNA samples to detect mutations using enzymes like CEL1, and identifying mutant individuals. TILLING has applications in functional genomics, genetic engineering, and evaluating genetic diversity. It provides a way to study gene function without transgenics.