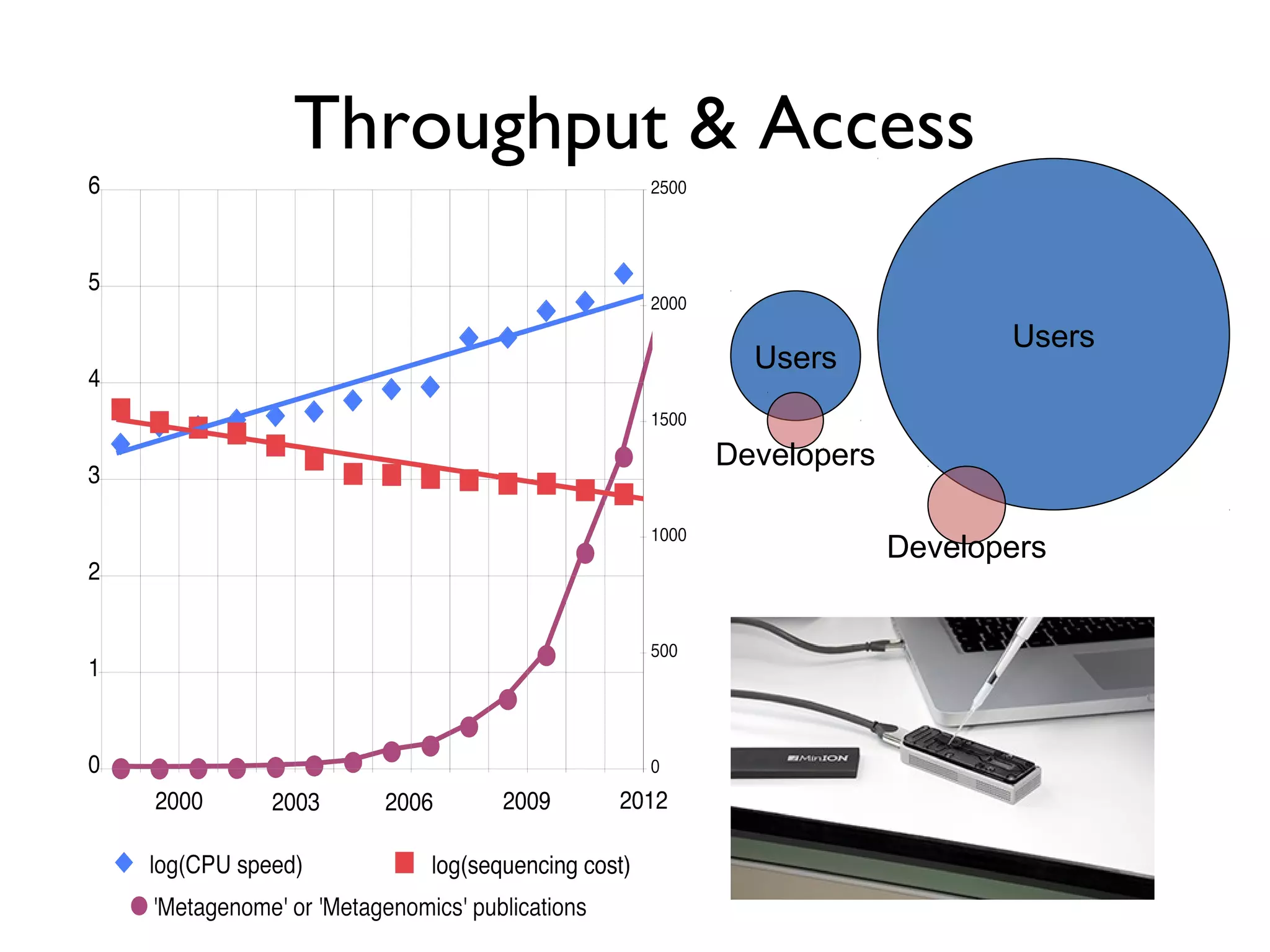
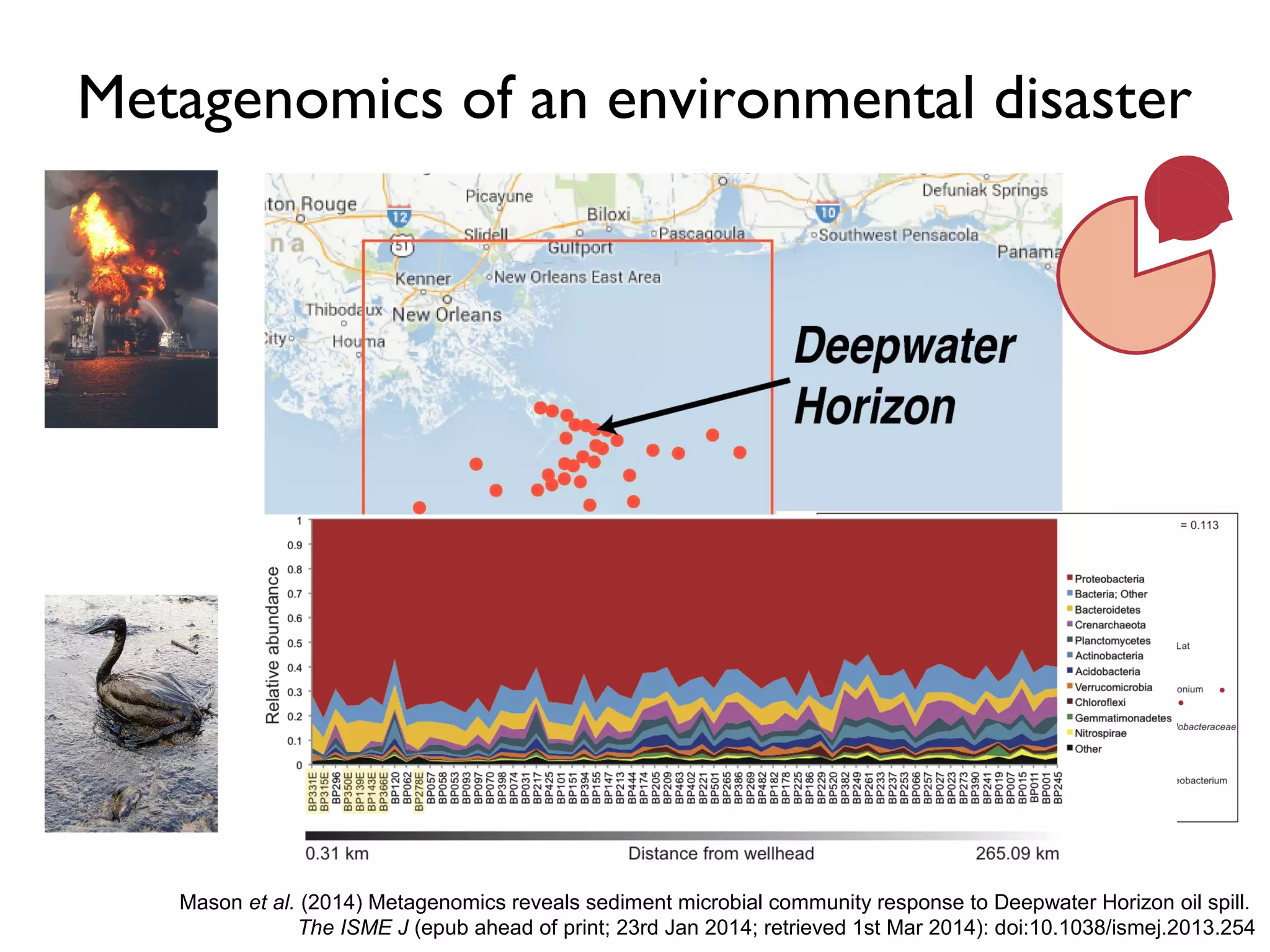
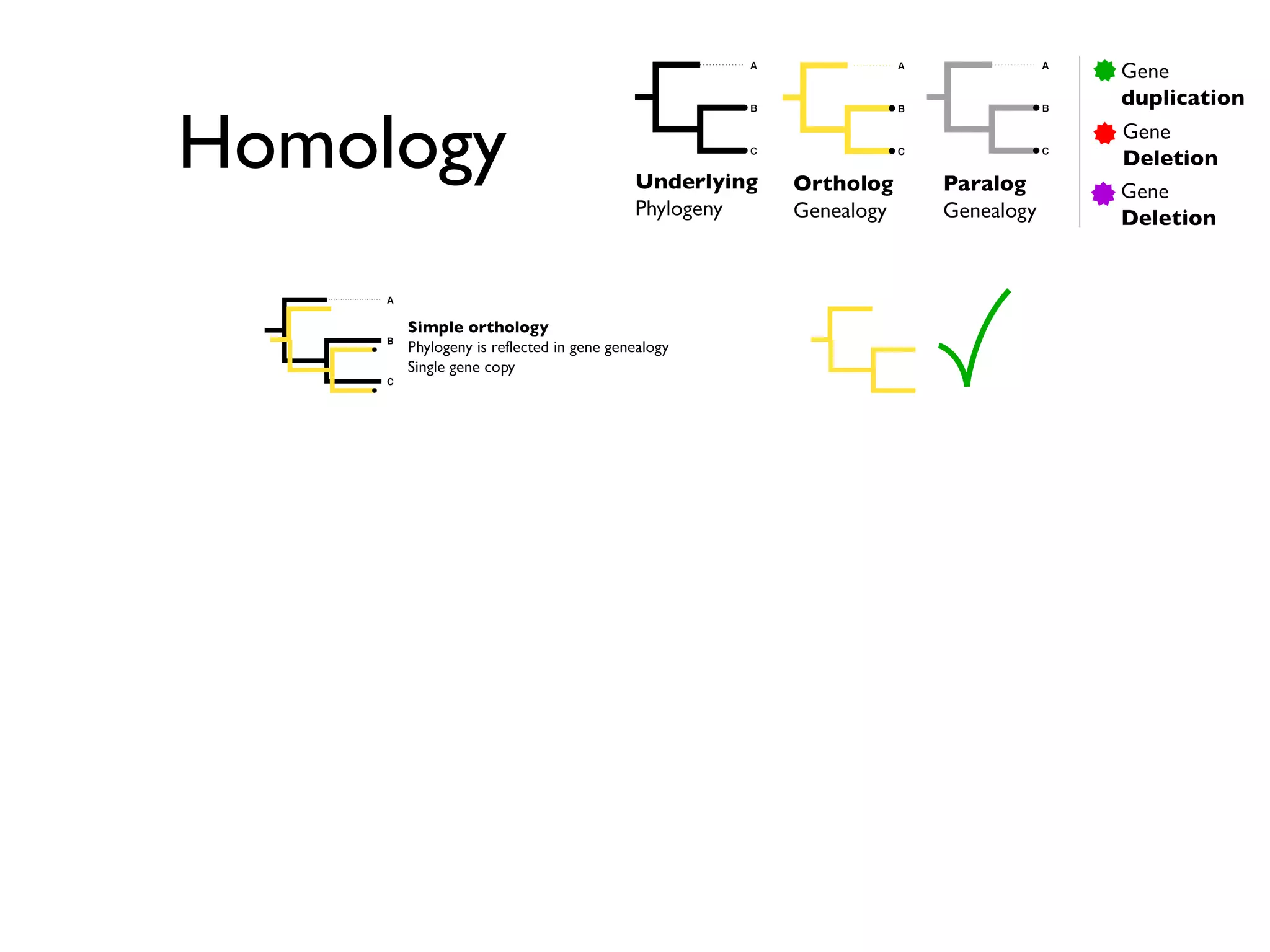
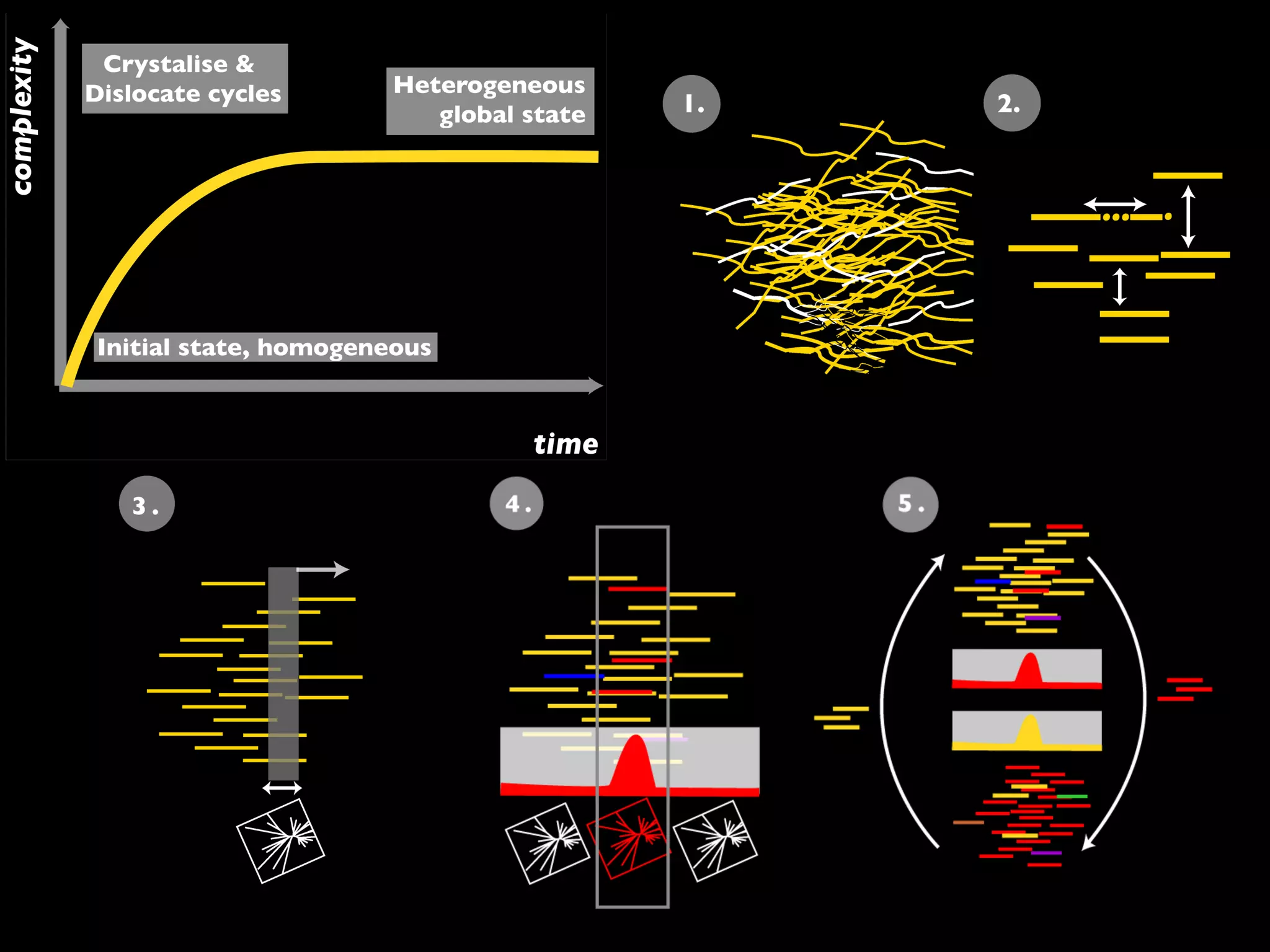
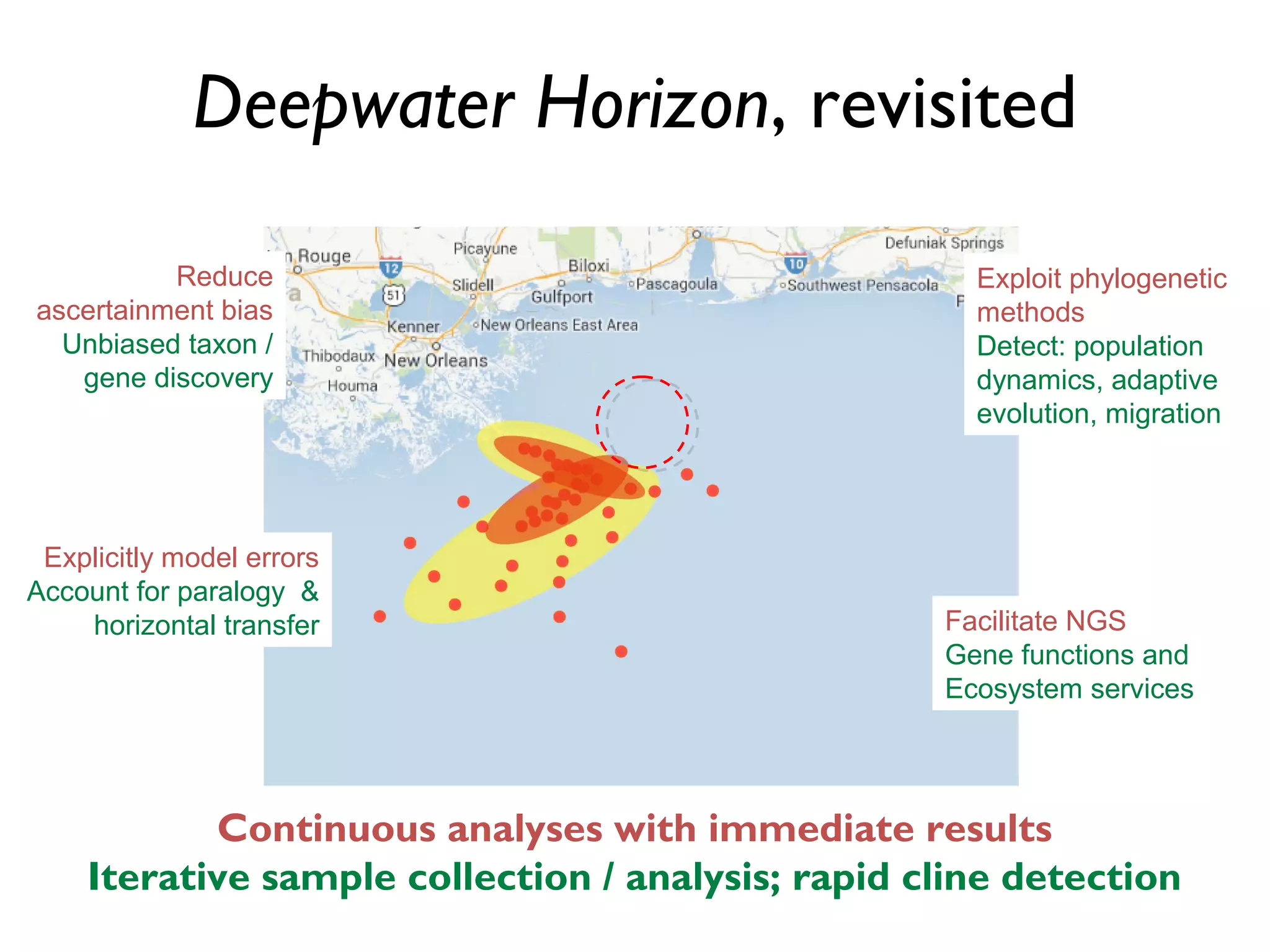
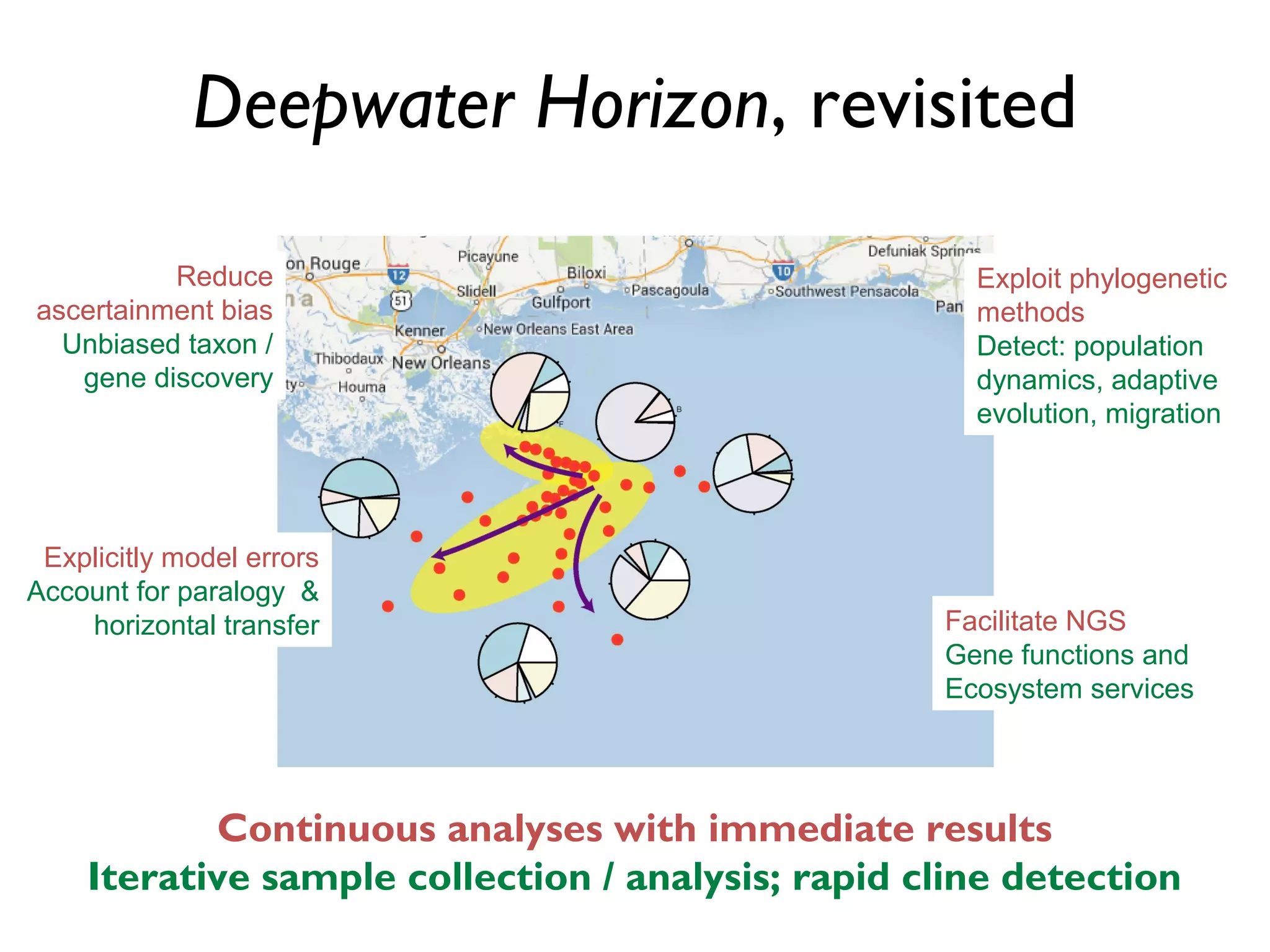
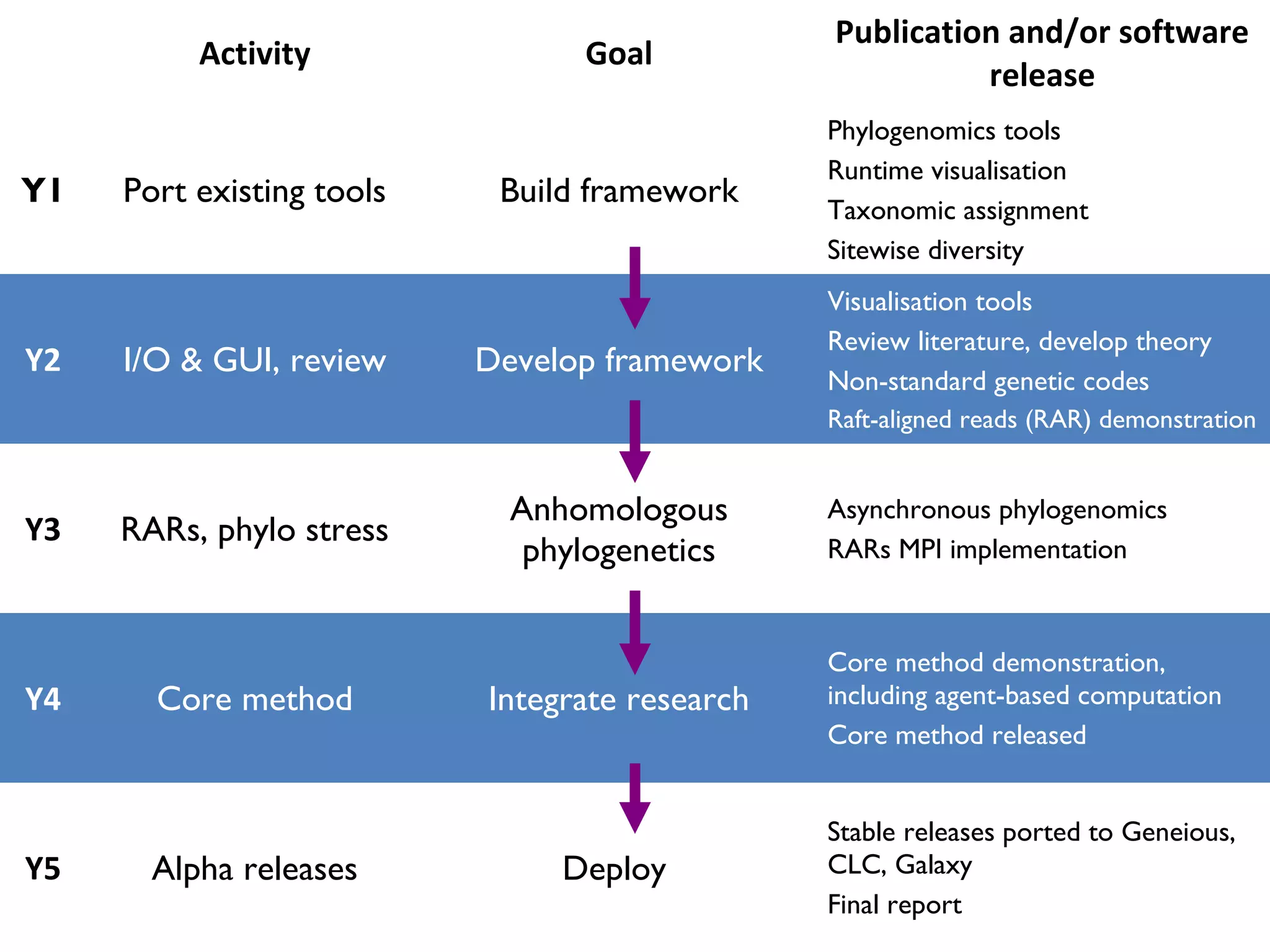
The document outlines a research project led by Joe Parker focused on advancing phylogenetic methods in environmental metagenomics, specifically in response to the Deepwater Horizon oil spill. It discusses the integration of high-throughput phylogenomics, machine-learning techniques, and various computing resources to improve taxonomic and genetic analyses. Key goals include the development of new software tools and frameworks to facilitate unbiased discovery of gene functions and evolutionary patterns.