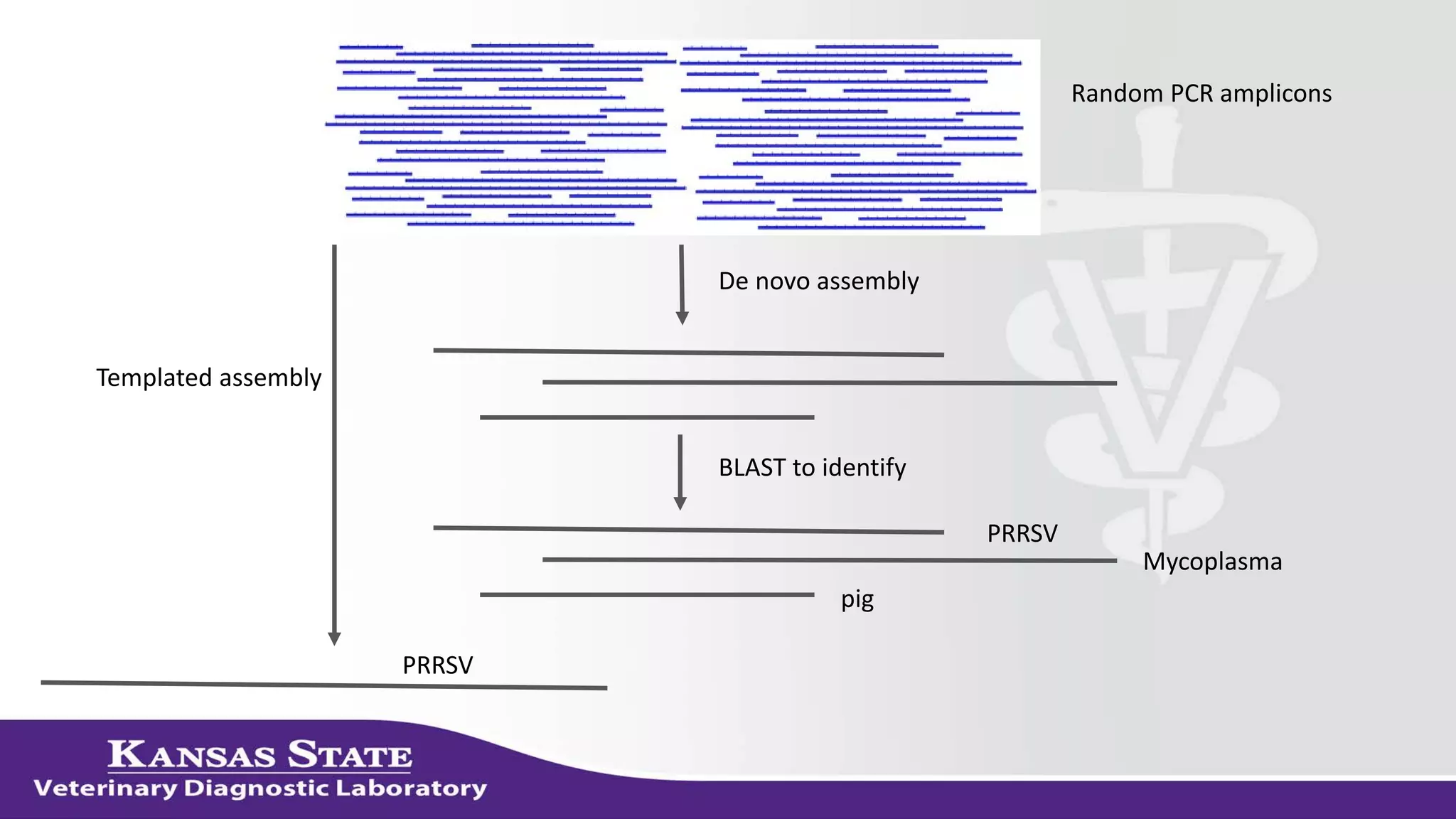
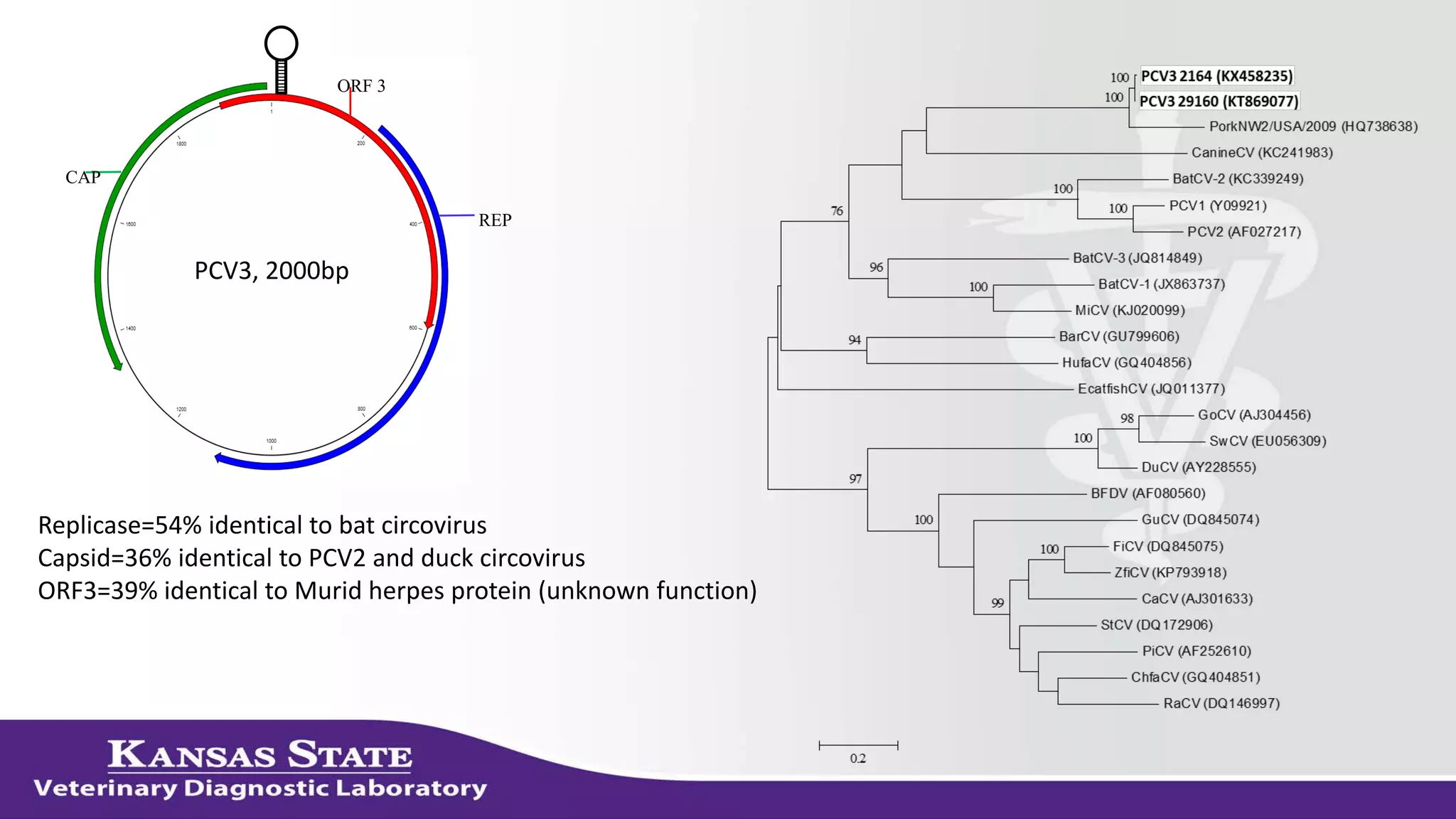
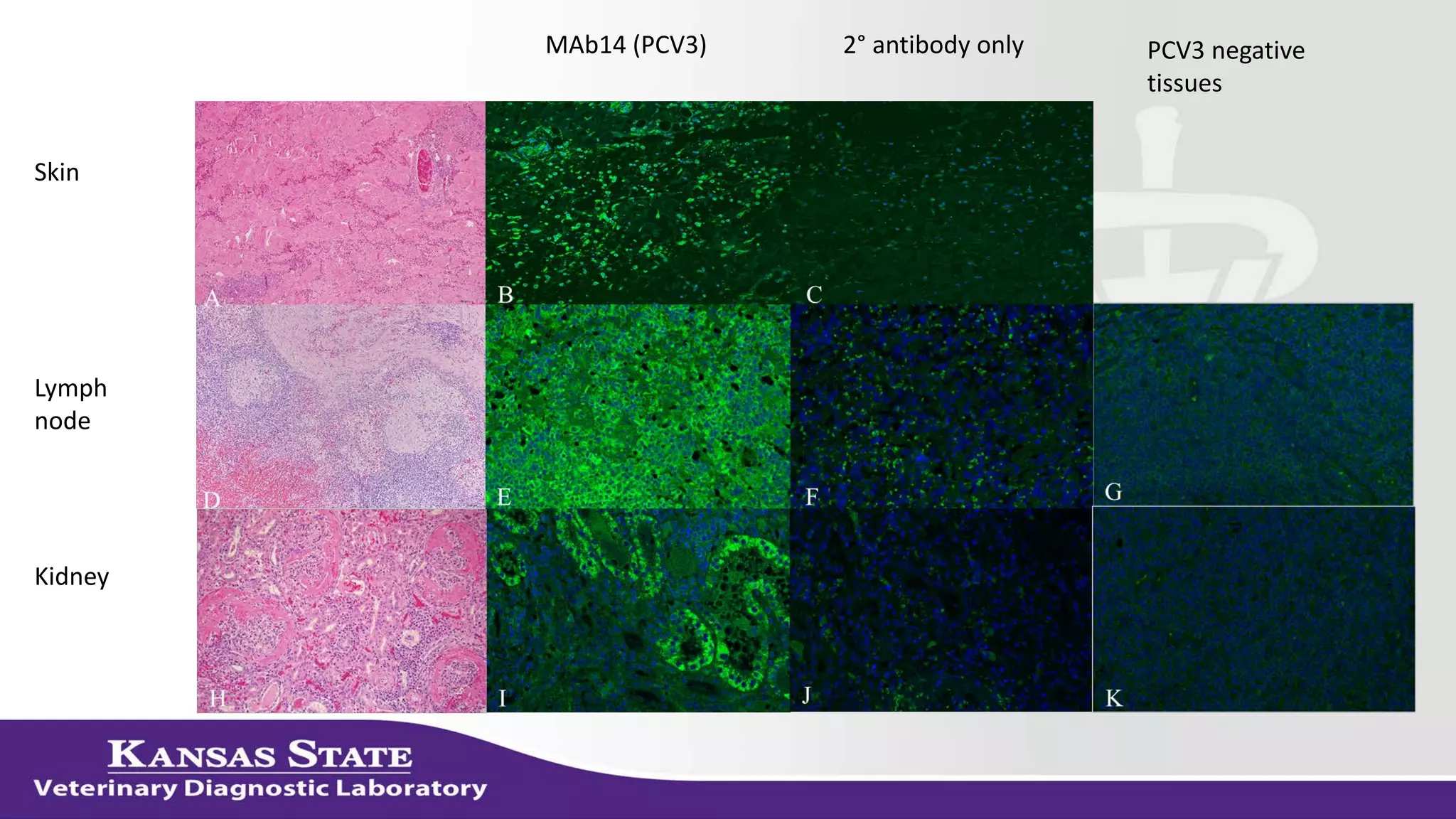
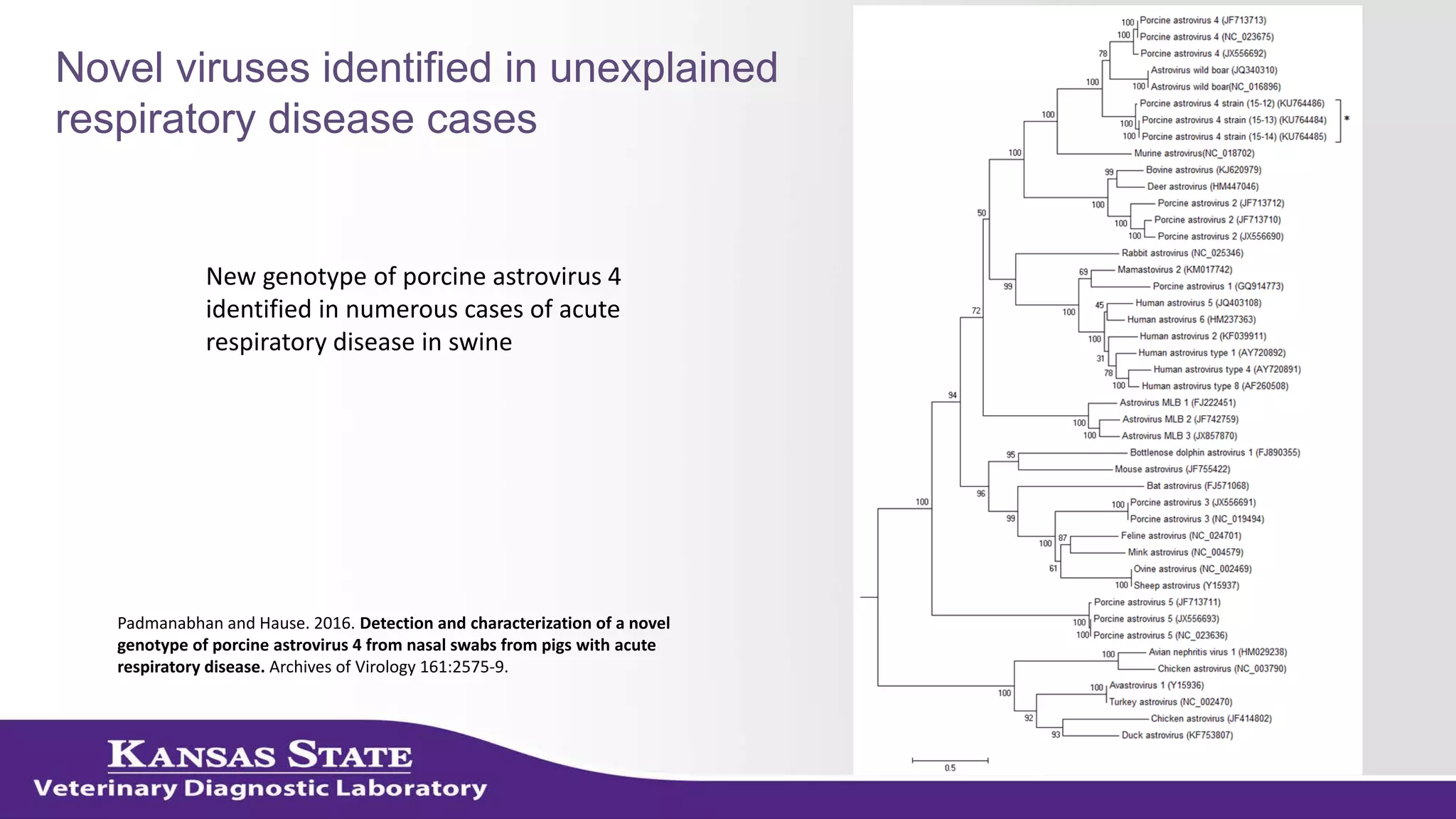
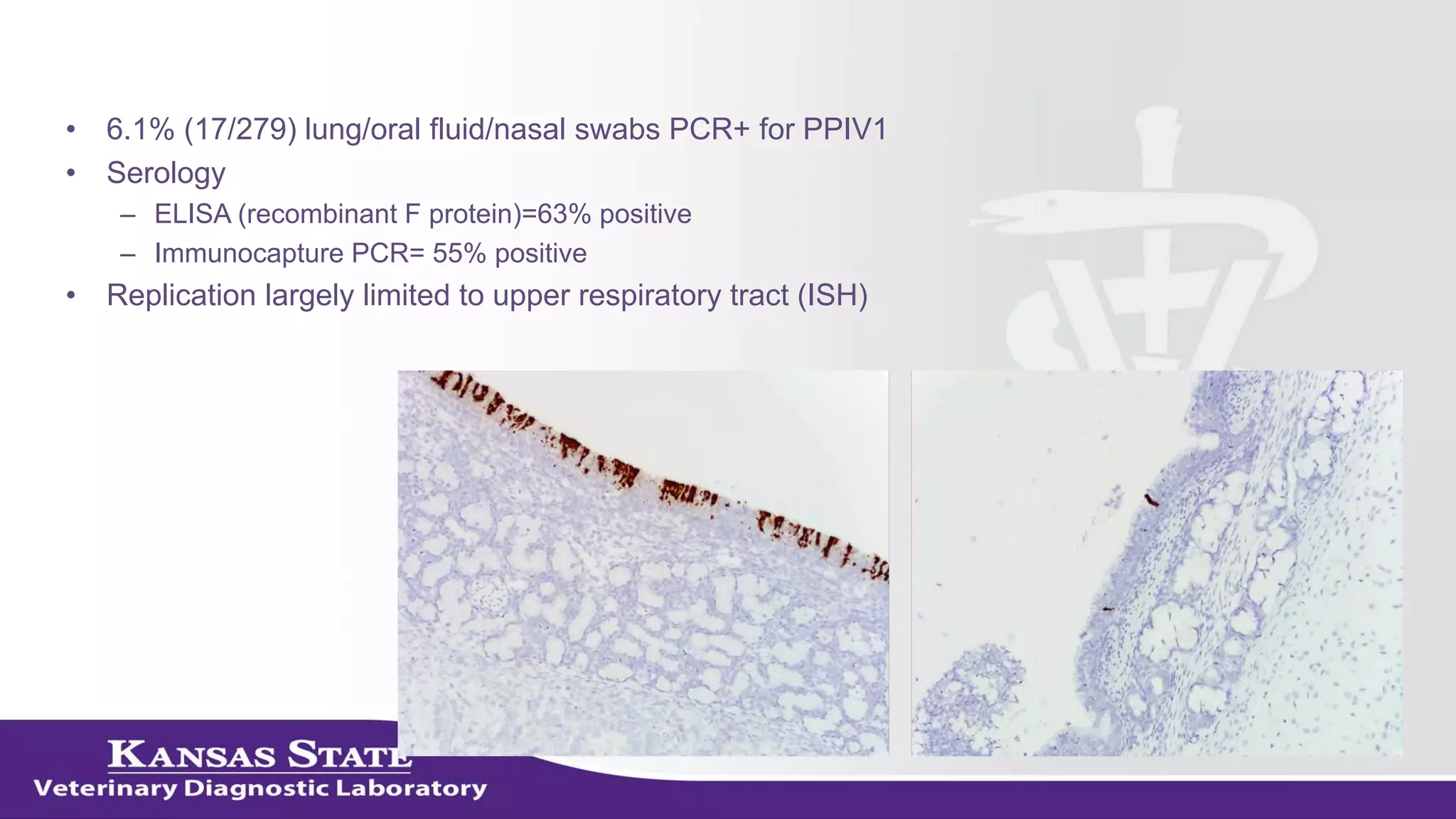
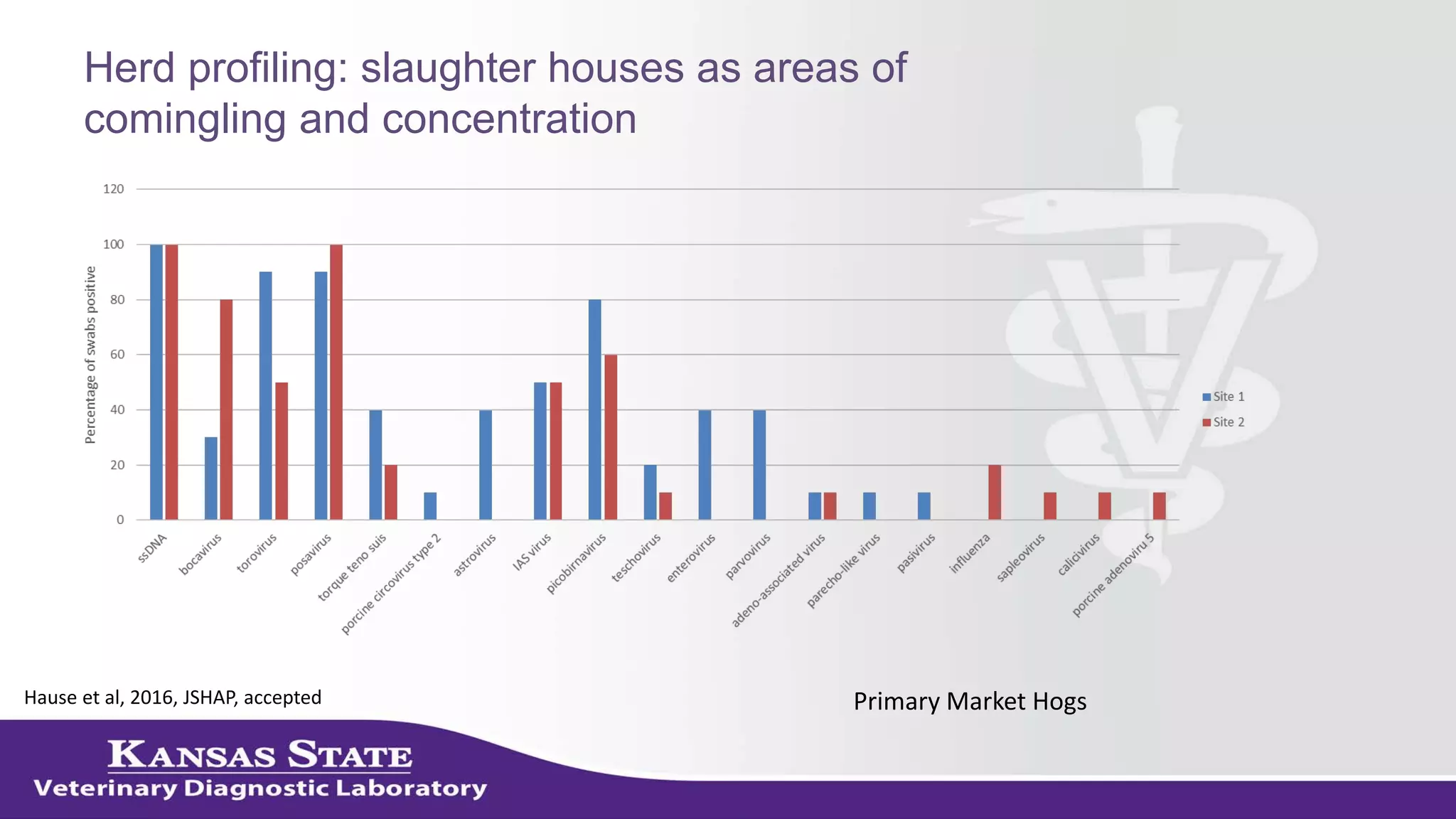
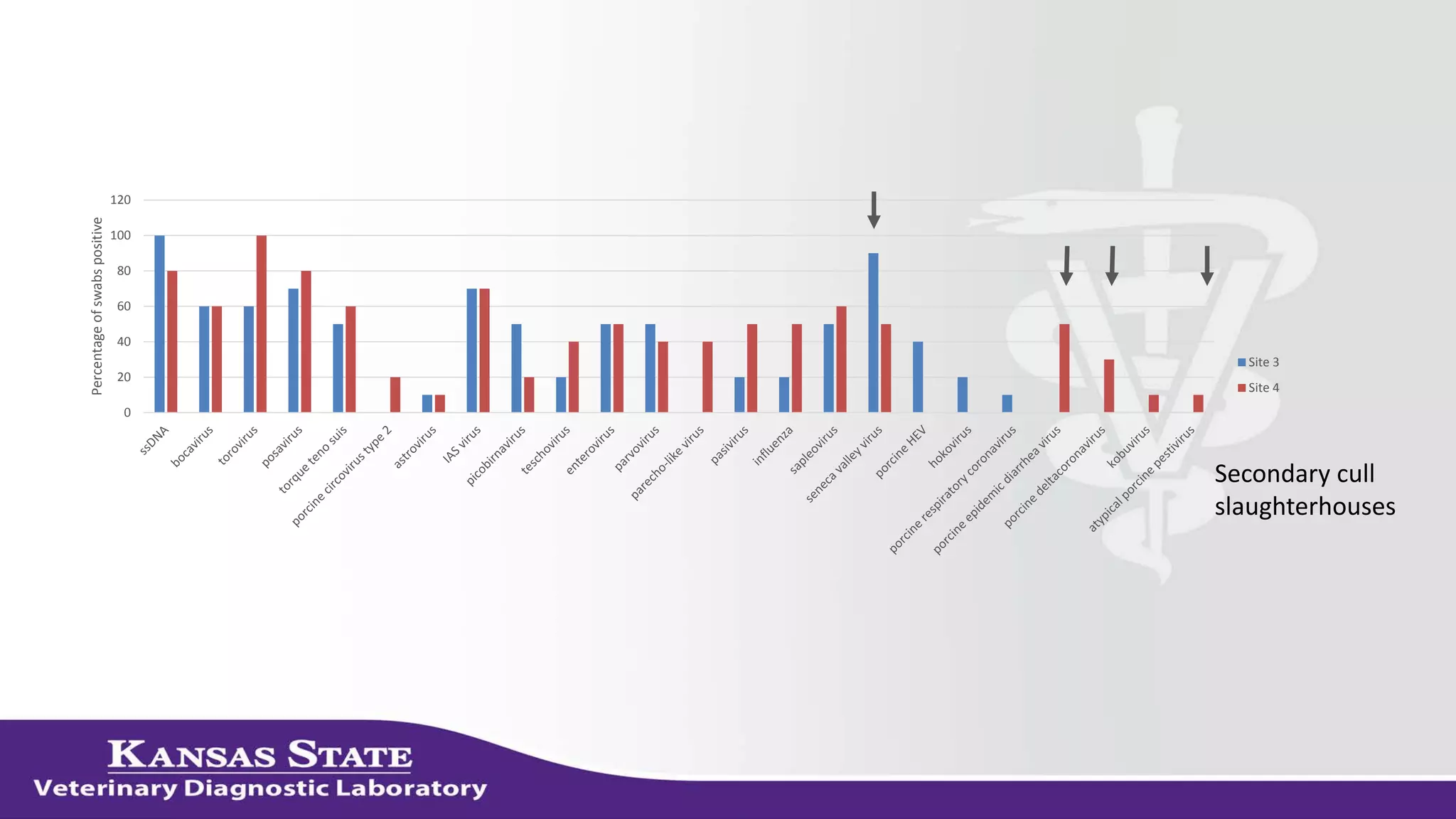
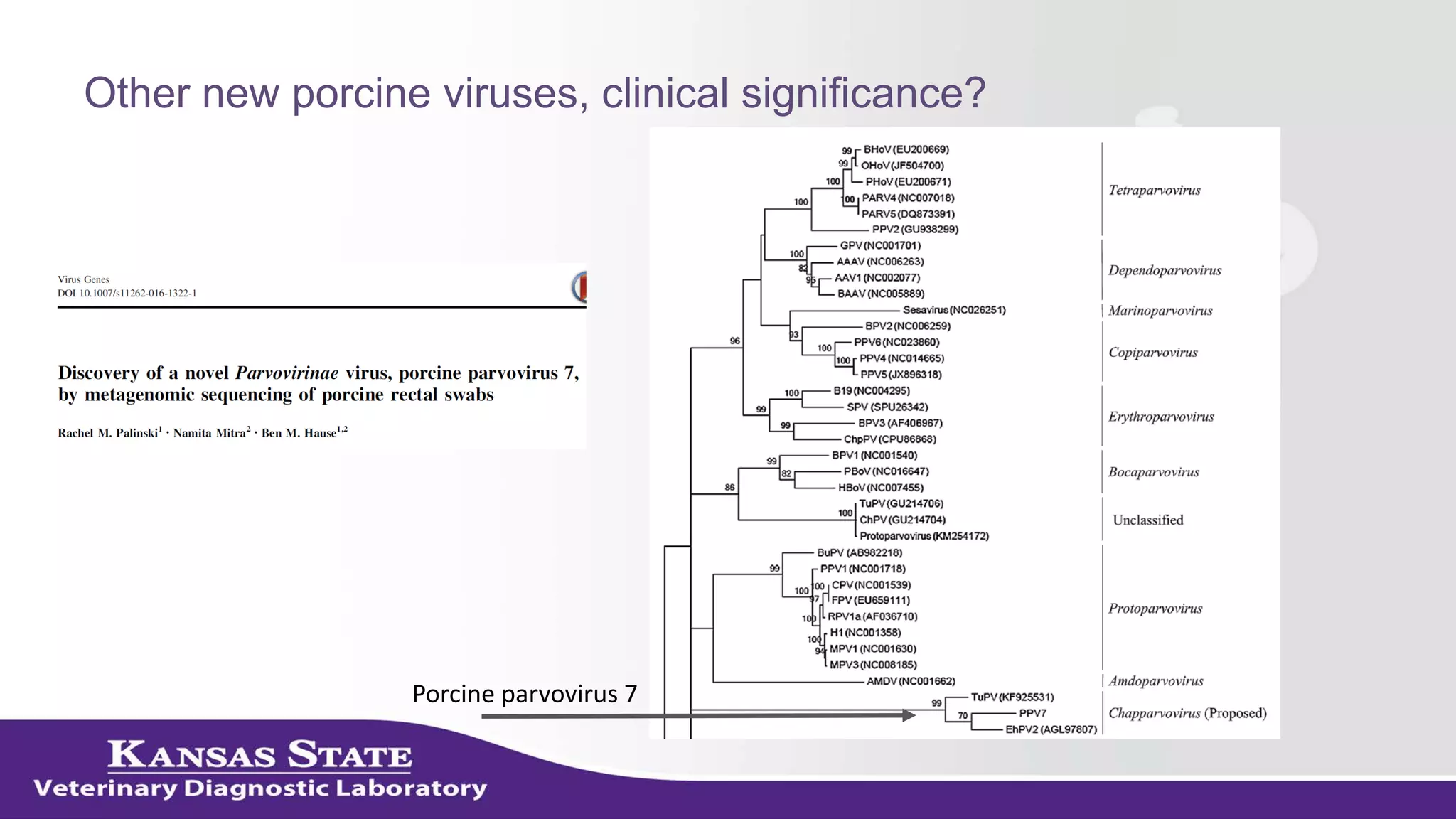
The document discusses the use of metagenomic sequencing for virus discovery in swine, detailing methods such as next-generation sequencing and including case studies on atypical porcine pestivirus and the newly identified porcine circovirus 3. It emphasizes the importance of metagenomic sequencing in diagnosing unusual clinical presentations and herd profiling, particularly in instances where conventional diagnostics fail. Additionally, it highlights the need for further research into the clinical significance of newly discovered porcine viruses.