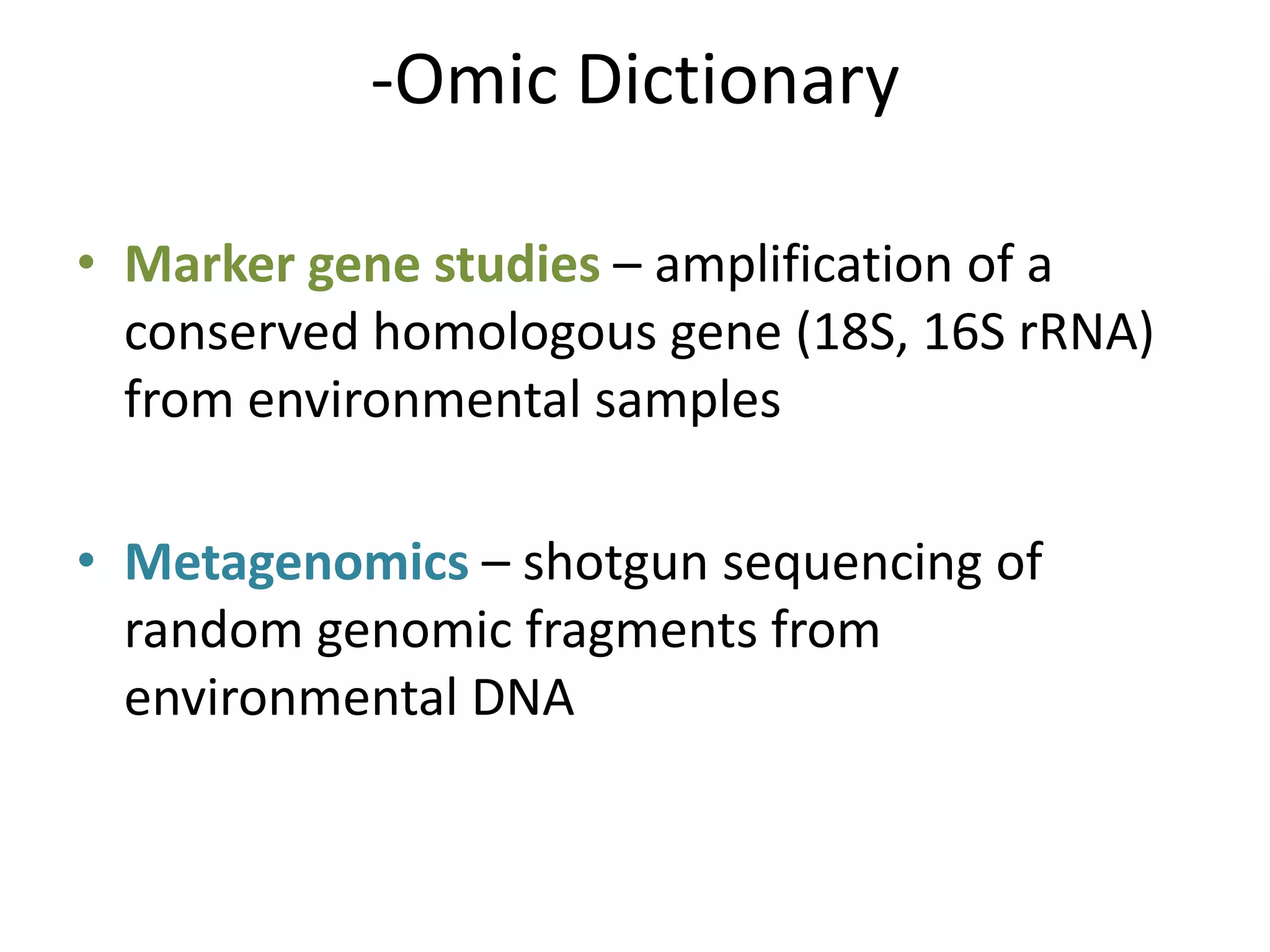
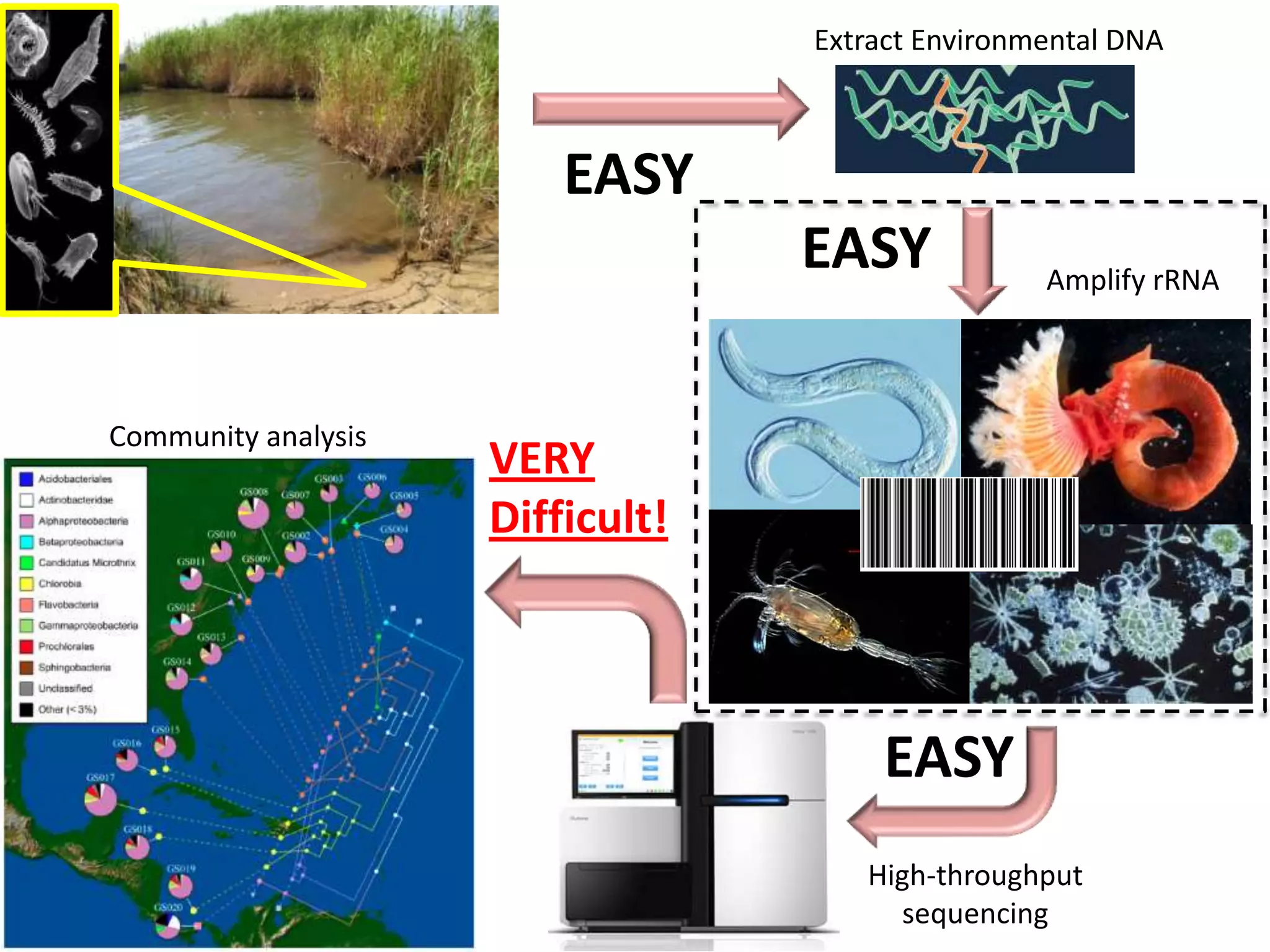
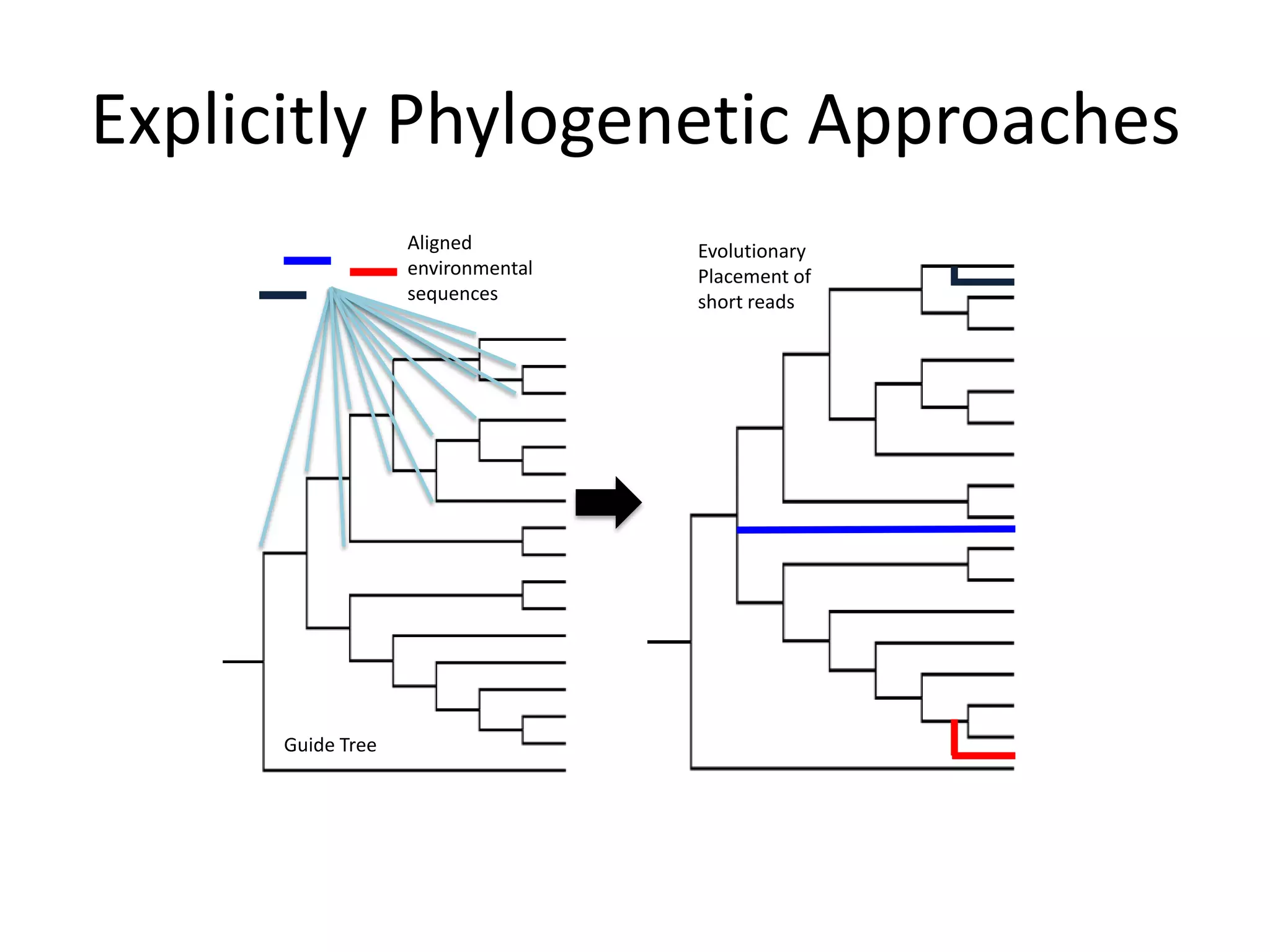
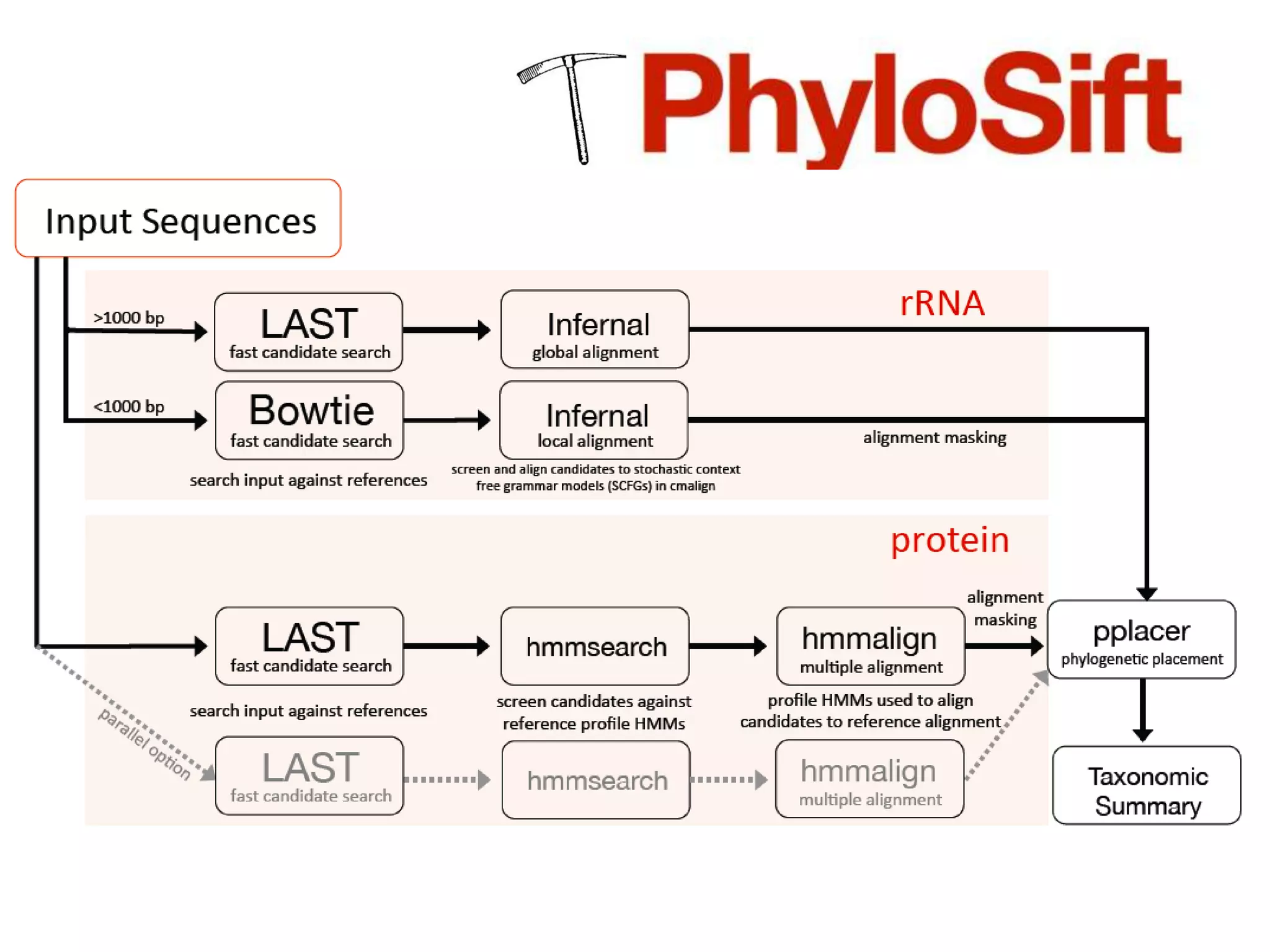
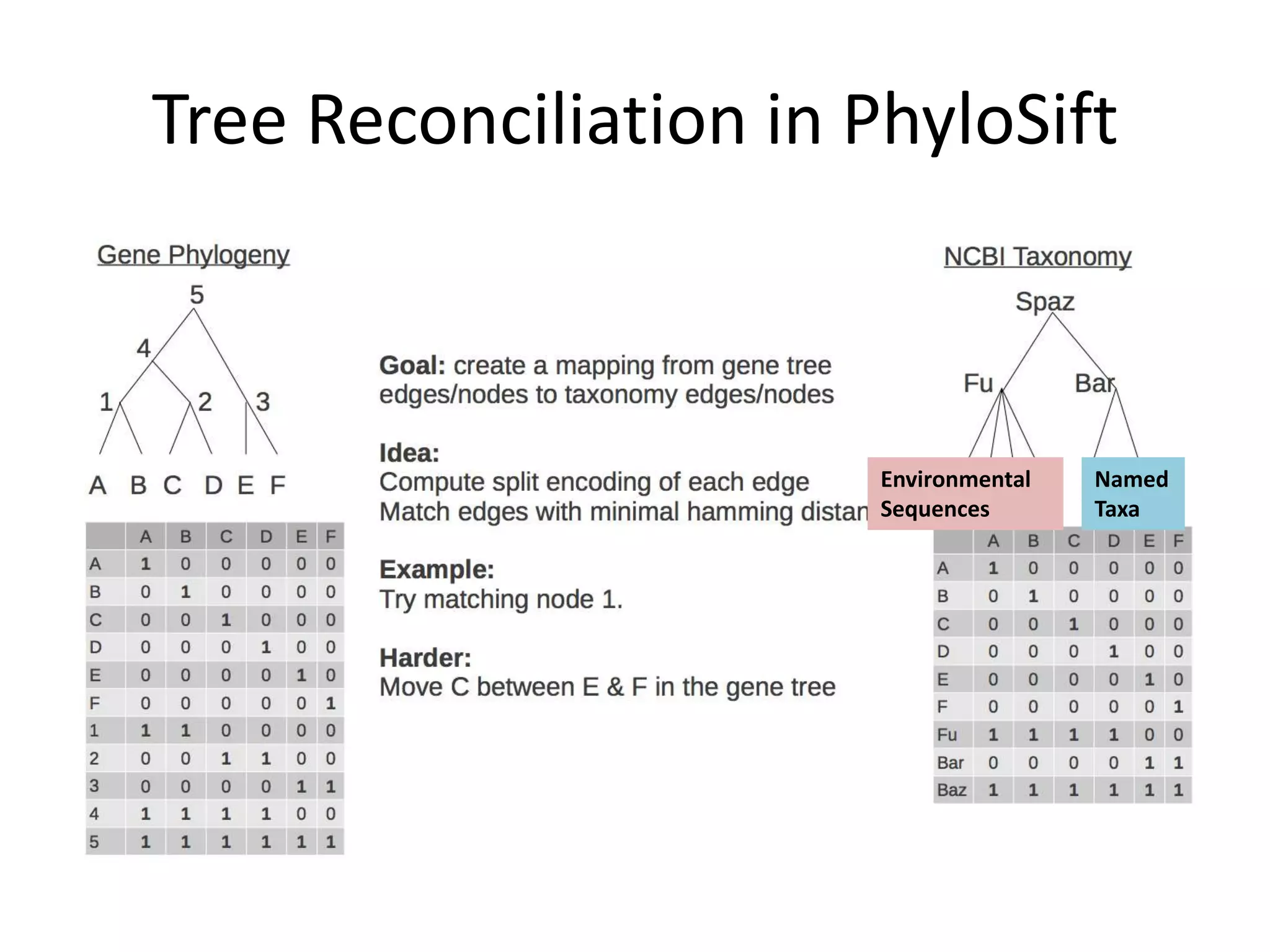
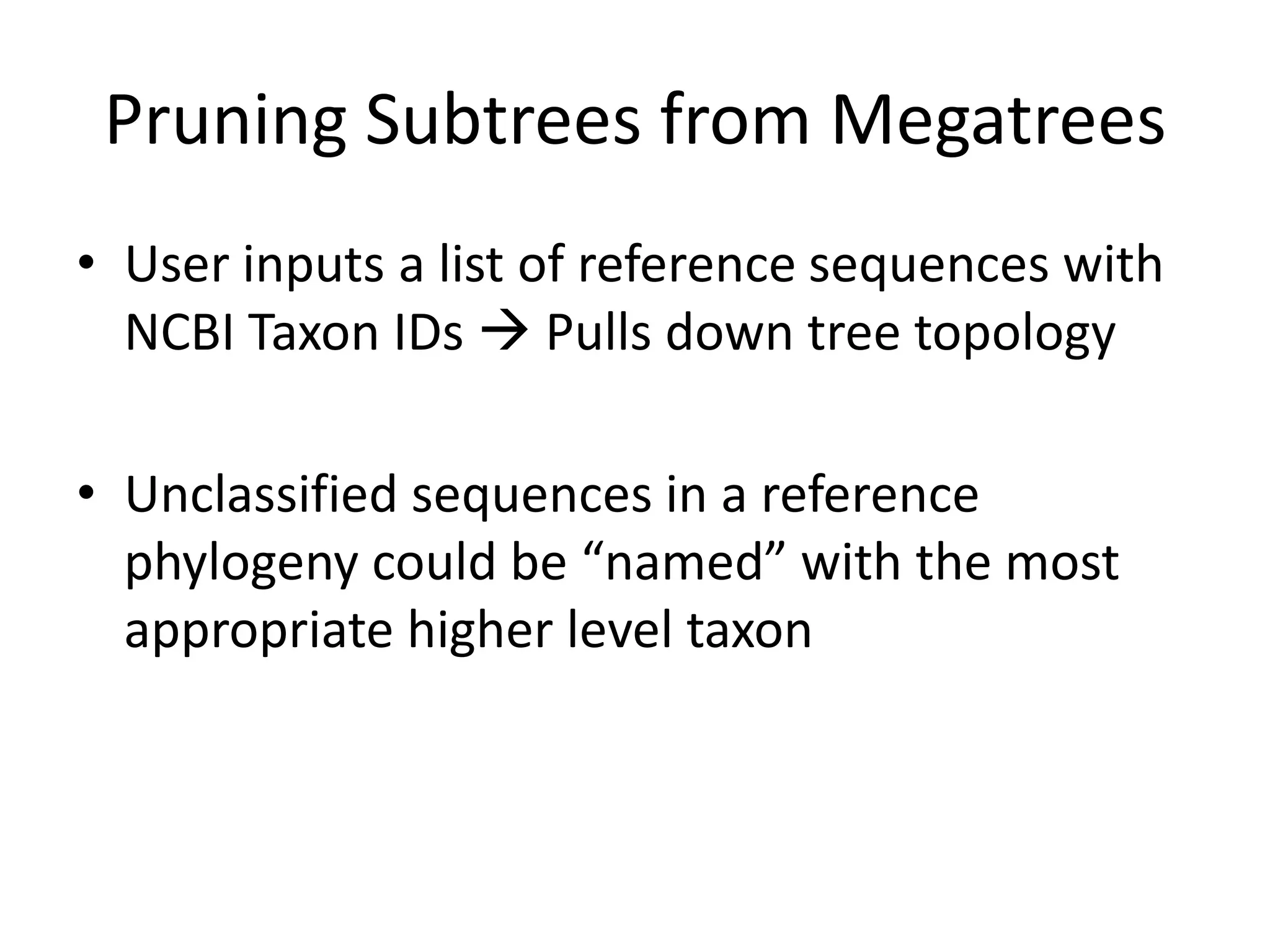
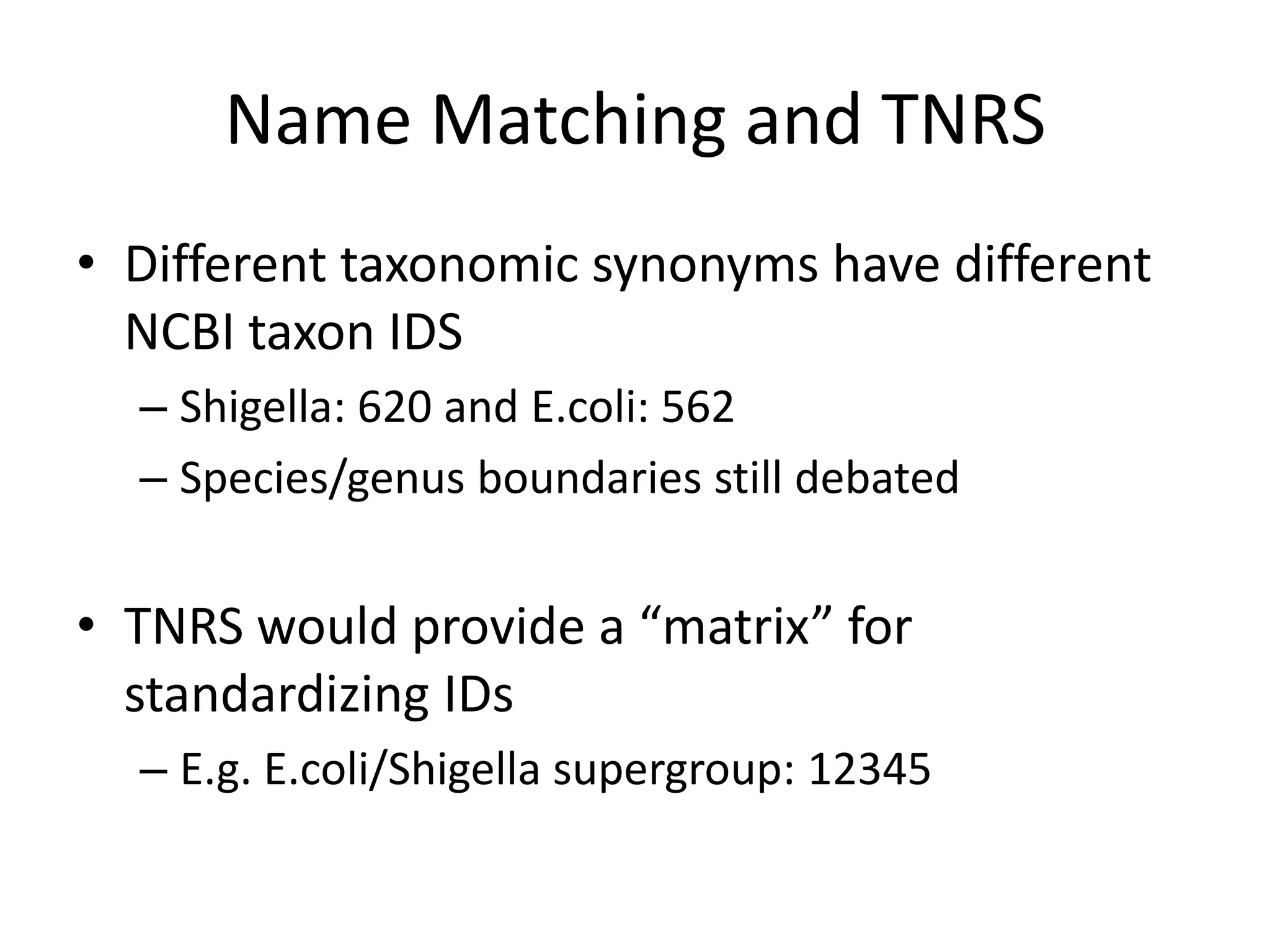
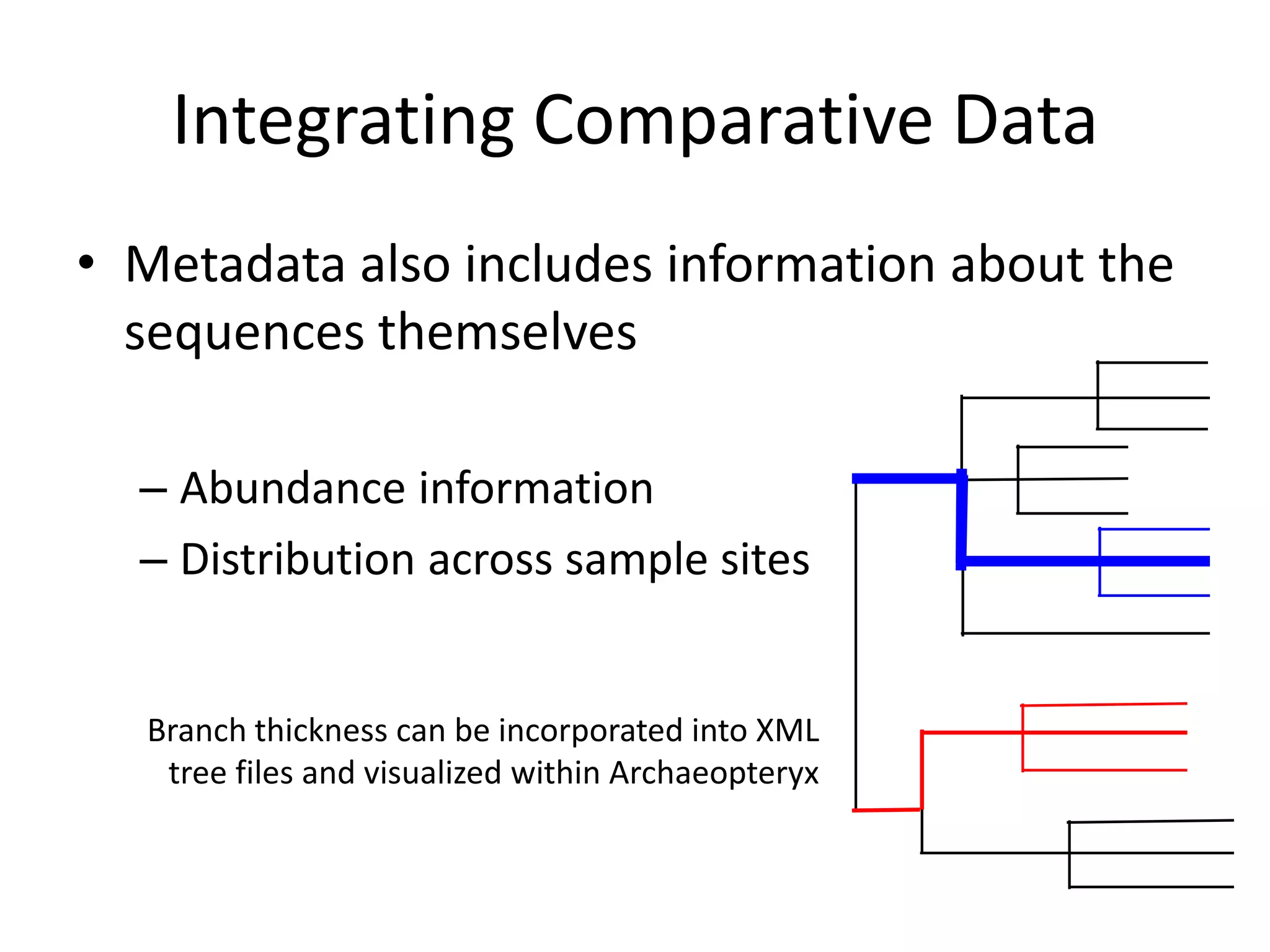
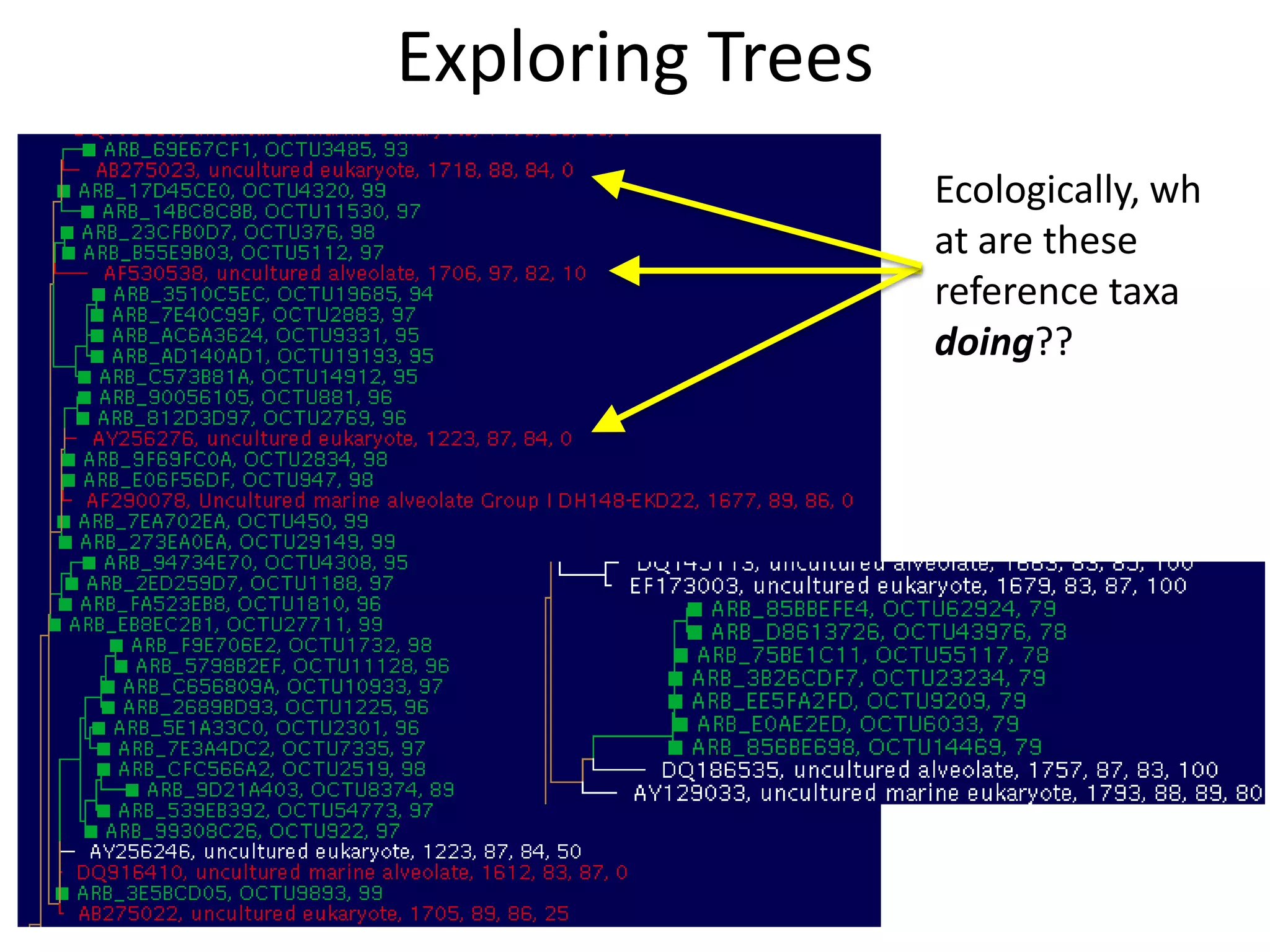
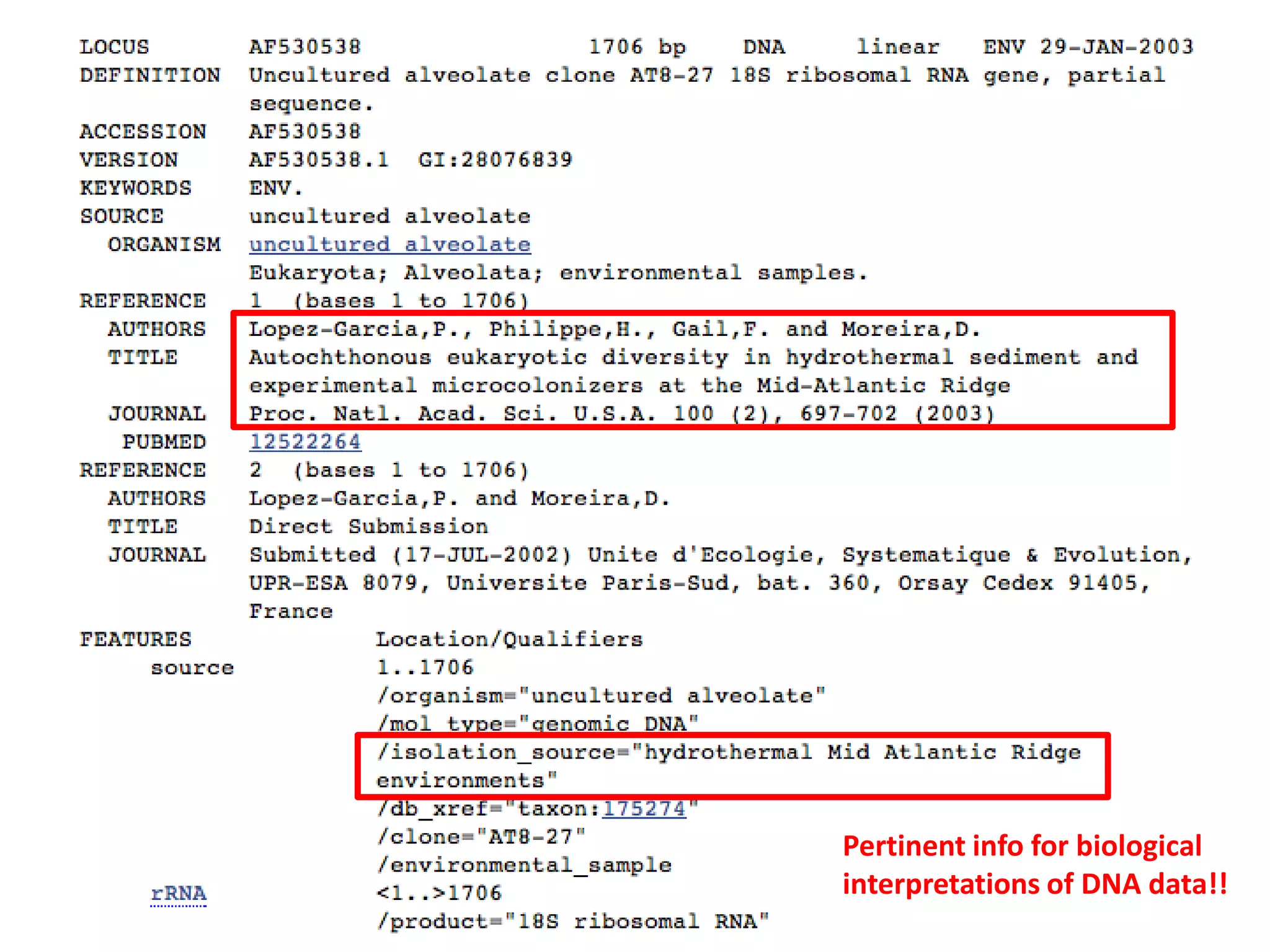
The document discusses the use of metagenomics and marker gene studies in analyzing environmental DNA, particularly through amplification of rRNA genes and high-throughput sequencing. It highlights the challenges in biodiversity and community analysis, emphasizing the integration of metadata for comparative data and ecological associations. Additionally, it outlines the process of using phylogenetic approaches for named sequences and the importance of standardizing taxonomic identifiers.