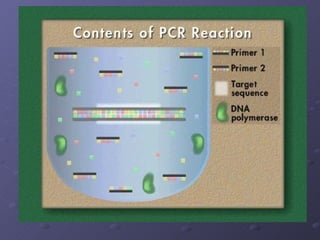
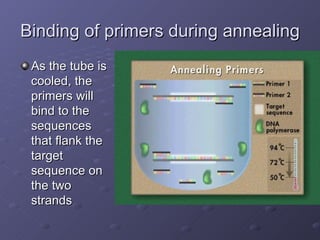
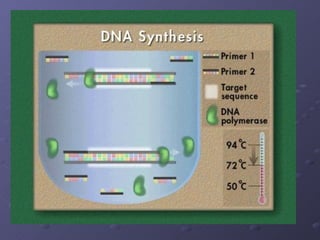
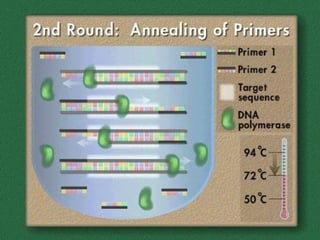
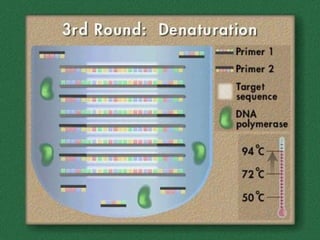
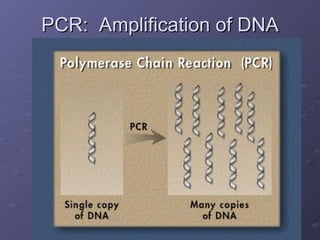
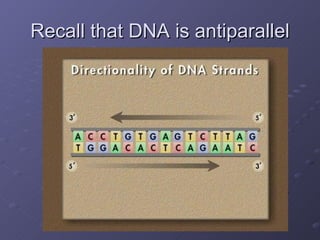
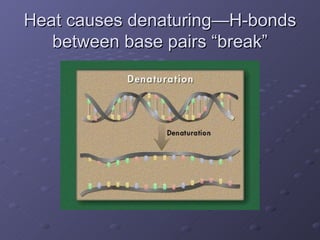
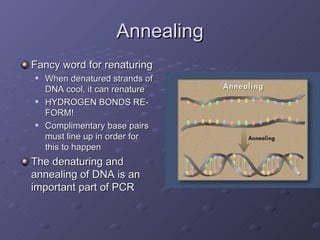
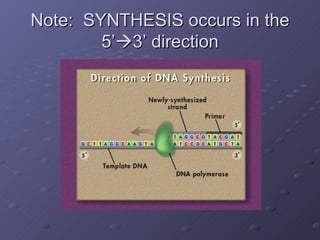
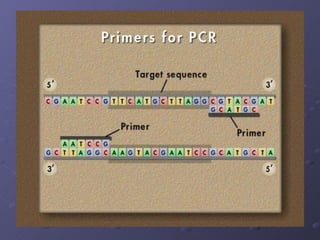
Polymerase Chain Reaction (PCR) allows scientists to make millions of copies of a specific DNA sequence starting with only a small sample. PCR involves repeated cycles of heating and cooling DNA to separate the strands and allow new strands to be synthesized using DNA polymerase. Kary Mullis invented PCR in 1983 while working at Cetus Corporation, and it has since become a fundamental technique for amplifying DNA sequences, enabling applications like DNA fingerprinting using very small forensic samples.





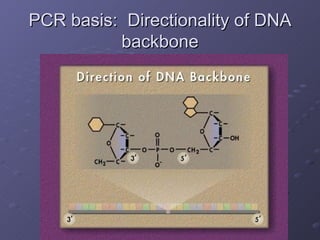





![PCR: Essential Reagents An excess of spare nucleotides An excess of primers We want the primers to bind to the sample DNA once it is denatured to prevent the strands from reannealing PLENTY of DNA polymerase [Taq polymerase in particular] Sample template of DNA](https://image.slidesharecdn.com/polymerase-chain-reaction-1228787797258056-8/85/Polymerase-Chain-Reaction-18-320.jpg)