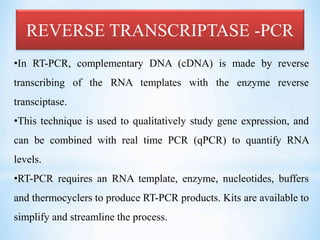
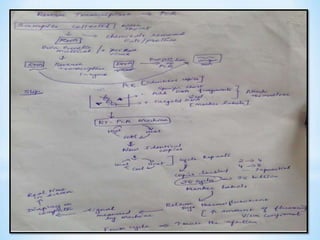
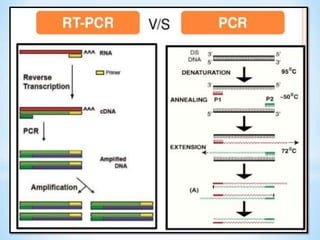
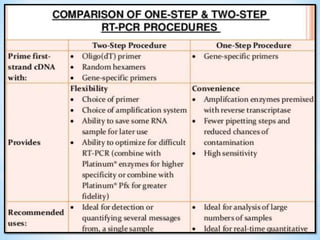
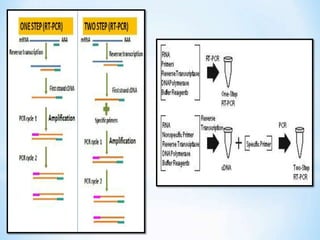
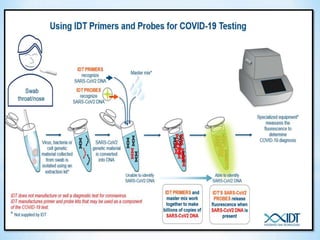
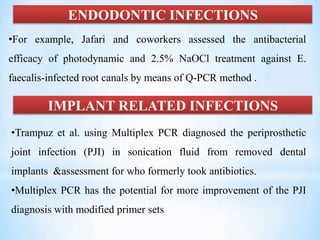
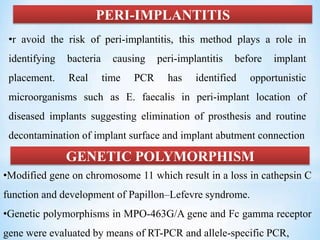
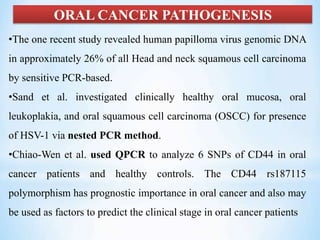
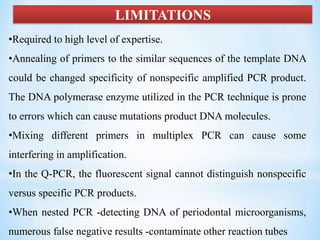
The document discusses polymerase chain reaction (PCR), including its history, basic requirements, essential components, principles, types, and dental applications. PCR is a technique used to amplify specific DNA sequences, allowing for large quantities of target DNA to be generated. It requires a DNA template, primers, DNA polymerase, and thermal cycling. Applications of PCR in dentistry include detecting viruses associated with periodontal disease and quantifying bacteria involved in dental caries.







































![REFERENCES
•Textbook of biochemistry – U satyanarayana
•N. Topcuoglu, G. Kulekci, 16S rRNA based microarray analysis of
ten periodontal bacteria in patients with different forms of
periodontitis, Anaerobe 35 (2015) 35-P.S. Anand, K.P. Kamath, S.
Anil, Role of dental plaque, saliva and periodontal disease in World
journal of gastroenterology: WJG 20(19) (2014) 5639.
• S. Tomo, HPV-16 DNA detection in fresh tissue, saliva and plasma
of patients with oral leukoplakia by real time PCR, (2018).
•L. Domingues, PCR, Springer2017. [16] G. Schochetman, C.-Y.
Ou, W.K. Jones, Polymerase chain reaction, The Journal of
infectious diseases 158(6) (1988) 1154-1157.](https://image.slidesharecdn.com/seminar21pcr-prinicple-201220163619/85/PCR-PRINCIPLES-64-320.jpg)
