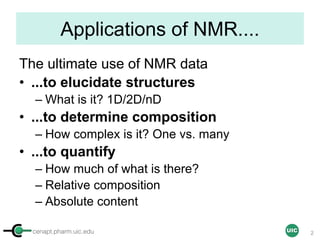
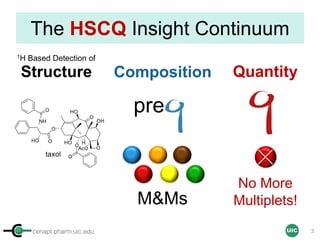
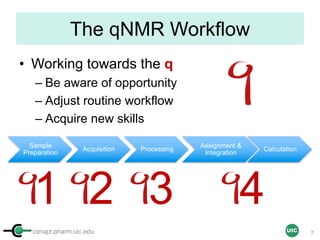
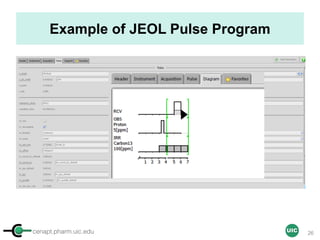
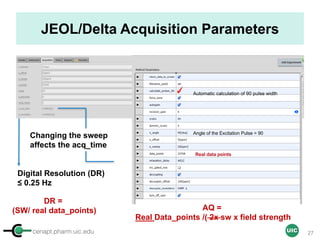
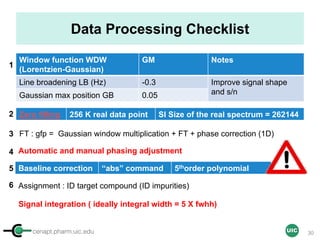
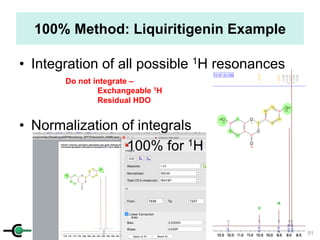
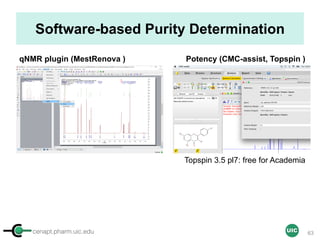
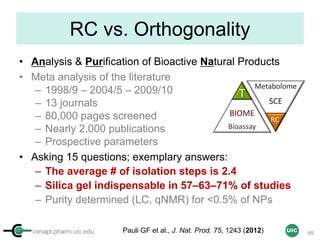
The document discusses the application of quantitative NMR (qNMR) for purity determination beyond mere structure elucidation. It outlines the goals of a workshop focused on qNMR, emphasizing the importance of accurate sample preparation, acquisition, and data processing as well as providing methodologies for impurity identification and purity calculation. Key details include the choice of qNMR methods, the use of internal and external calibrants, and parameter optimization for reliable quantitative analysis.






![cenapt.pharm.uic.edu UIC
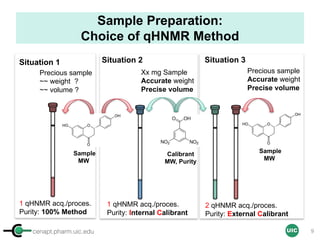
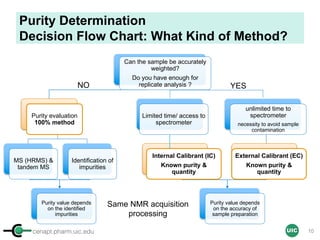
Sample Preparation with IC
(Example 1 = Several Precious Samples)
Sample
Calibrant (IC)
STEP 5STEP 1
Qty
[IC] ≈ [Sample]
stock solution
(commercial)
STEP 2 STEP 3
STEP 4
ID, MW
ID, MW,
Purity
Qty
Max 500 µL (5 mm)
Max 200 µL (3 mm)
Tared vial
+ dried cpd
15](https://image.slidesharecdn.com/03handsonpracticev08website-170814170234/85/qHNMR-for-purity-determination-15-320.jpg)
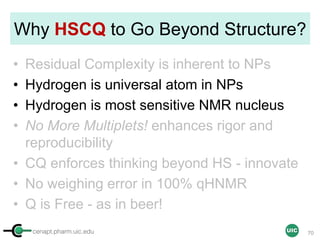
![cenapt.pharm.uic.edu UIC
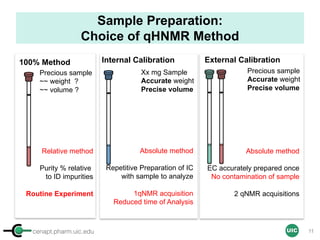
Sample Preparation with IC
(Example 2 = Non-precious Powder Sample)
Sample
STEP 4STEP 1 STEP 3
ID, MW
ID, MW, Purity, [IC]
[IC] ≈ [Sample]
In mM
OR
Commercial IC
stock solution
Adjust the concentration
STEP 2
Calibrant
Qty
Max 500 µL (5 mm)
Max 200 µL (3 mm)
16](https://image.slidesharecdn.com/03handsonpracticev08website-170814170234/85/qHNMR-for-purity-determination-16-320.jpg)
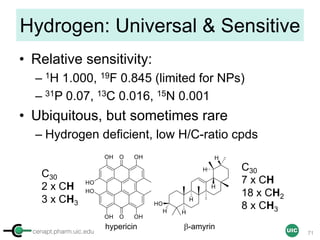
![cenapt.pharm.uic.edu UIC
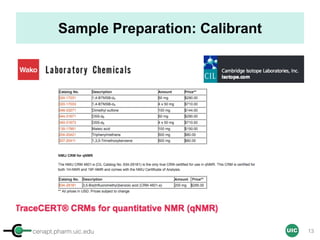
Sample Preparation with EC
Sample
STEP 1 STEP 2
ID, MW
ID, MW, Purity, [EC]
[EC] ∝ [Sample]
In mM
OR
Commercial solution
Calibrant
Qty Max 500 µL (5 mm)
Max 200 µL (3 mm)
STEP 3 STEP 4 STEP 4
17](https://image.slidesharecdn.com/03handsonpracticev08website-170814170234/85/qHNMR-for-purity-determination-17-320.jpg)



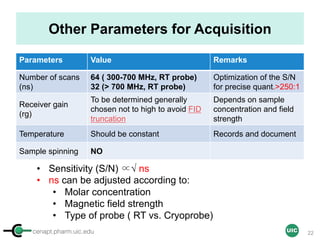
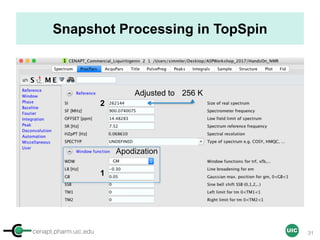
![cenapt.pharm.uic.edu UIC
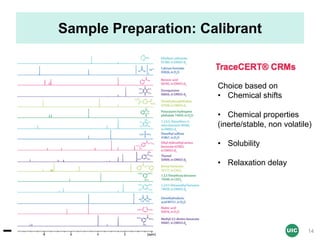
Key Parameters for Acquisition
Parameters Value Remarks
Relaxation delay
(D1)
Theory: 5 X T1 of compounds
Practice: 60 sec (if T1 unknown)
Accuracy of the integral
values
Pulse width
(90 pulse: P1)
to be determined using the 360
null
Function of solvent
nucleus, sample
Acquisition time
(AQ)
4 sec
Optimization of digital
resolution
Spectral window
(SW)
transmitter offset
(O1P)
SW: 30 ppm – O1P:7.5 ppm
SW: 20 ppm – O1P: 4.5 ppm
For an optimal baseline
correction (edge
effects), and digital
resolution
FID size (TD) 32 K-64 K For better resolution
AQ = (TD/[2x SW])
Number of Data Points (TD) = 2 x Spectral Width (SW in ppm X field strength) x Acquisition Time (AQ)
21](https://image.slidesharecdn.com/03handsonpracticev08website-170814170234/85/qHNMR-for-purity-determination-21-320.jpg)
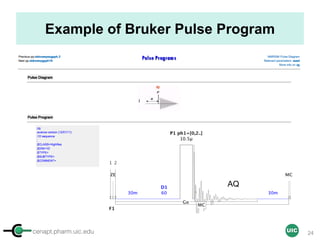

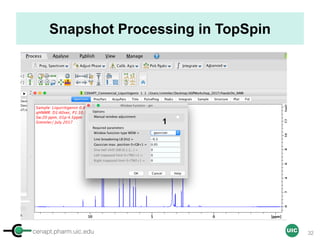
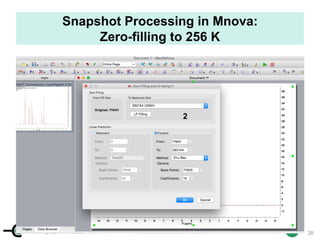
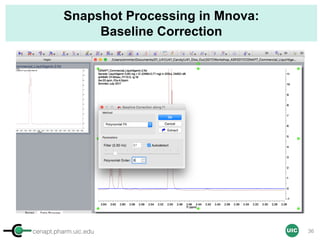
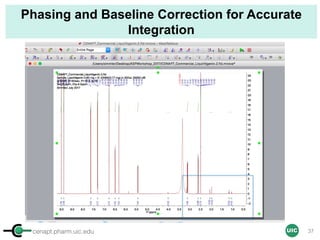
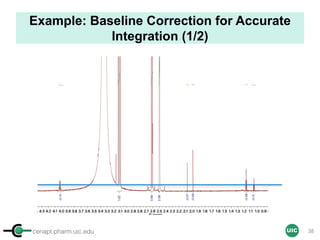
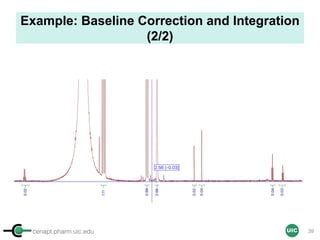

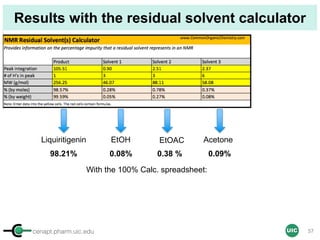
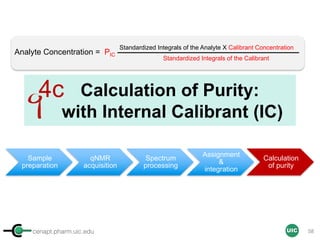
![cenapt.pharm.uic.edu UIC
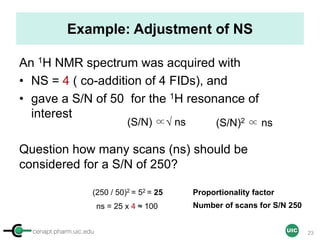
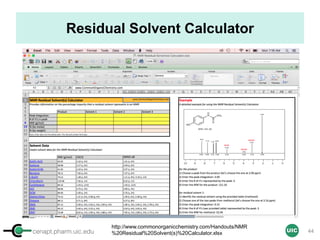
Calculation of Purity
Pauli et al. Journal of Medicinal Chemistry 57, 9220–31 (2014)
Concentration (molarity) α [integral Area/ Number of nuclei]
1 qNMR experiment
• ID compound
• ID impurities
• Residual solvent
• Moisture analysis
• Purity
4b
45](https://image.slidesharecdn.com/03handsonpracticev08website-170814170234/85/qHNMR-for-purity-determination-45-320.jpg)
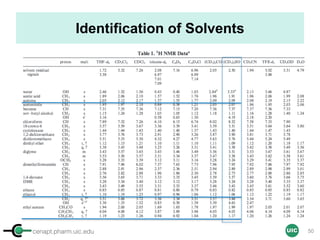
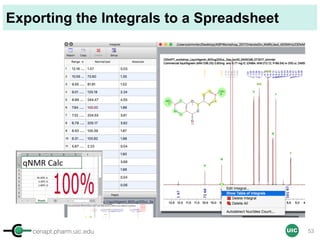

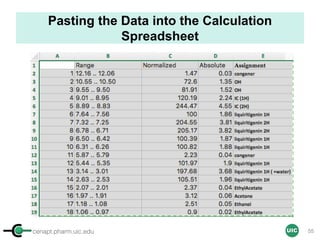
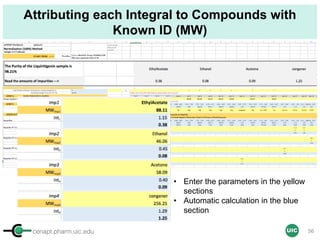
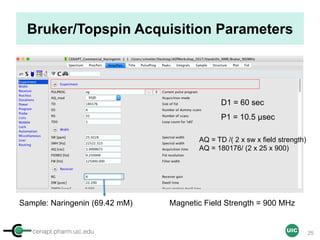
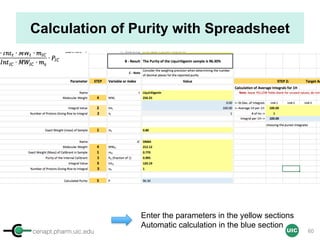
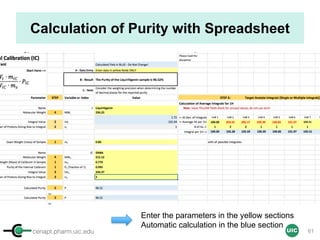
![cenapt.pharm.uic.edu UIC
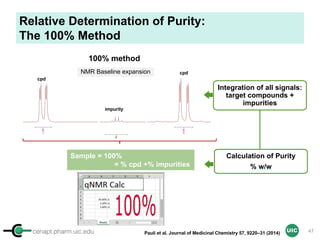
Purity Determination with IC: Overview
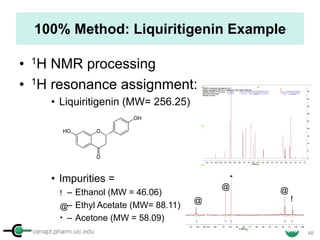
Sample: Liquiritigenin
MW : 256.25
Qty: 0.80 mg
Calibrant (IC)
MW: 212.12
Qty: 0.77 mg
Purity:99.54
IC : 1H
target : 1H
𝑃 [%] =
𝑛𝐼𝐶 ∙ 𝐼𝑛𝑡𝑡 ∙ 𝑀𝑊𝑡 ∙ 𝑚𝐼𝐶
𝑛𝑡 ∙ 𝐼𝑛𝑡𝐼𝐶 ∙ 𝑀𝑊𝐼𝐶 ∙ 𝑚 𝑠
∙ 𝑃𝐼𝐶 =
1 x 100
t = target compound = liquiritigenin
256.25 0.77
99.54
1 x 120.19 212.12 0.80x x
x x
= 96.30% w/w
59
Qty (mic) = n/MW → Intic
→ InttTrue Qty (mt) = n/MW??](https://image.slidesharecdn.com/03handsonpracticev08website-170814170234/85/qHNMR-for-purity-determination-59-320.jpg)
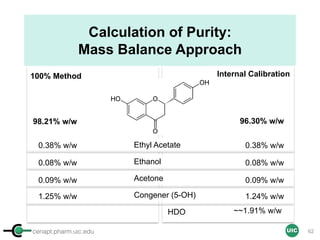



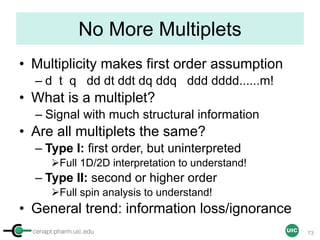





![cenapt.pharm.uic.edu UIC
Purity Activity-Relationship (PAR)
• Is ursolic acid (MIC 32-128 µg/mL) a viable anti-TB lead?
– Eight UA accessions: declared vs. found 1-16 % differences
• qNMR Answer: Inverse correlation between purity and activity
• qNMR Net Outcome Pure UA is essentially inactive
82
O
OH
CH3
CH3
CH3
CH3
CH3
HO
H
H
H
1
4 6
9
10
12
19
14
20
17
24 23
25
26
27
28
29
30
CH3H3C
Purity [%]
65 70 75 80 85 90 95 100 105
Anti-TBMIC[ug/ml]
0
50
100
150
200
250
300
SI=IC50/MIC[ug/ml]
0.0
0.1
0.2
0.3
0.4
0.5
IC50VERO
0
5
10
15
20
25
30
% purity vs MIC H37Rv
% purity vs MIC H37gfp
% purity vs SI
% purity vs IC50 VERO
Jaki et al. J. Nat. Prod. 71, 1742-8 (2008)](https://image.slidesharecdn.com/03handsonpracticev08website-170814170234/85/qHNMR-for-purity-determination-82-320.jpg)
