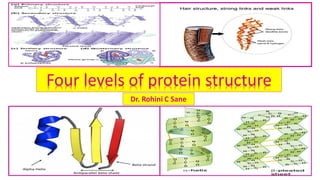
Four levels of protein structure
- 1. Four levels of protein structure Dr. Rohini C Sane
- 2. Structure of proteins ❖ Proteins are polymers of amino acids and made up of one or more polypeptide chains . ❖ Every protein in its native state has a unique three dimensional structure which is referred to as its conformation. ❖The number and sequence of these amino acids in the protein are different in different proteins. ❖The function of a protein arises from its conformation. ❖Protein structure can be classified into four levels of organization.
- 3. Configuration and Conformation of a molecule Configuration of compound denotes the spatial relationship between particular atoms e.g. L- amino acids and D-amino acids Conformation of molecule means the spatial relationship of every atom in a molecule e.g. rotation of a portion of the molecule
- 4. Four levels of protein structure Proteins are the polymers of L--amino acids. The structure of proteins is rather complex which can be divided into four levels of organization. Proteins: polypeptide with more than 50 amino acid residues
- 5. Four levels of structural organization of proteins ❖ Proteins are polymers of amino acids and made up of one or more polypeptide chains . ❖Four levels of structural organization can be recognized in proteins: 1. Primary structure: is determined by the number and sequence of amino acids in the protein. 2. Secondary Structures: is the conformation of polypeptide chain formed by twisting or folding . It occurs when amino acids are linked by hydrogen bonds to form -helix and -sheets . 3. Tertiary Structure : is the three dimensional arrangement of protein structure. It is formed when alpha-helices and beta-sheets are held together by weak interactions. 4. Quaternary structure: occurs in protein(oligomers) consisting of more than one polypeptide chain where certain polypeptides aggregate to form one functional protein.
- 6. Four orders of protein Structure Primary structure is determined by the sequence of amino acids. Secondary Structures: the number and sequence of amino acids in the protein. It occur when amino acids are linked by hydrogen bonds to form -helix and -sheets . Tertiary Structure : is the three- dimensional arrangement of protein structure. Quaternarystructure:occursinprotein(oligomers) consistingofmorethanonepolypeptidechainwherecertain polypeptidesaggregatetoformonefunctionalprotein.
- 7. Four orders of protein structure: Bricks Wall Rooms Building Amino acids (Bricks ) Secondary structure (bends /twist in walls) Tertiarystructure(self–containedroom) Primary structure (walls) Quaternarystructure(buildingwithsimilar/dissimilarrooms Structural Hierarchy of proteins
- 8. Structural hierarchy of proteins • Primary structure : is a linear sequence of amino acids forming a backbone of proteins .It refers to the order in which amino acids are linked together in the peptide chain. e.g. Glutathione: Tripeptide : Glutamic acid-Cysteine-Glycine Methionine Enkephalins : pentapeptide: Try- Gly- Gly- Phe -Met • (N-terminal end) → H2N - - COOH (C-terminal end) • Peptide bond → linear , planner ,rigid ,partial double bond character ❖Secondary Structures :spatial arrangement of proteins by twisting of polypeptide chain= folding patterns in proteins (alpha-helix ,beta-sheet ) ❖Tertiary Structure : Three dimensional structure generated by interaction between the amino acid residues of functional proteins. ❖Quaternary structure : refers to the spatial arrangement of subunits of proteins which are joined by non-covalent interactions . This is seen in proteins with two or more polypeptides chains(oligomers). ❖Super secondary Structures :indicate folding patterns in proteins
- 9. Primary structure of proteins • Primary structure of proteins denotes the number and sequence of amino acids in protein. The successive amino acids are linked by peptide bond(covalent bond). • Generally ,amino acids are arranged as a linear chain . Each component amino acid is called a residue or moiety. Very rarely, proteins may be a branched form or circular form (Gramicidin) . Primarystructureofproteinsislargelyresponsibleforitsfunctions. • The branching points in the polypeptide chain may be produced by interchain disulphide bridges (the covalent disulphide bonds between different polypeptide chains in the same protein) or portion of the same polypeptide chain (intrachain). They are part of primary structure. • Eachpolypeptidewillhaveanaminoterminal(N-terminal)withfreeaminogroupandacarboxyterminal ends(C-terminal)withfreecarboxygroup.By convention, they are represented with amino terminal on the left and carboxy terminal end on the right end. • The amino acids composition of protein determines its physical and chemical properties. • Primarystructureofproteins(sequenceofaminoacids)isdeterminedbythegenescontainedinDNA. Primarystructureofnumberofproteinsareknowntoday.e.g.Insulin, Glucagon,Ribonuclease,Growth hormones.AnychangeinthePrimarystructureofproteinsaffecttheirfunctions.
- 10. Peptide bond formation • Formation of peptide bond = a covalent bond is formed by amide linkage between the - carboxyl group of one amino acid and - amino group of another amino acid by removal of a water molecule. • Successive amino acids are joined/cemented by peptide bond ( -CO-NH) in proteins . ➢The peptide bonds form strong backbone of polypeptide and side chains of amino acid residue project outside the peptide backbone. Dipeptide :two amino acids and one peptide bond ( not two bonds)
- 11. Characteristics of Primary structure of proteins:1 1. Apeptidecontains twoormoreaminoacidresiduesjoinedtogetherbyapeptidebonds.Individualamino acidscanbeconsideredasbricks. 2. Formationofpeptidebond=acovalentbondisformedbyamidelinkagebetweenthe-aminogroupof oneaminoacidand-carboxylgroupofanotheraminoacidbyremovalofawatermolecule. 3. CharacteristicsofaPeptidebond: a. Rigid,covalent,stable,strongandcanbehydrolyzed bytheactionofproteolyticenzymes,acidsand alkalis b. Planerandwithpartialdoublebond(nofreedomofrotation)incharacter c. –C=O,NH–Existintrans-configuration.Bothgroupsarepolarandinvolvedinhydrogenbonds. d. Impartstabilitytotheprimarystructureofproteins(disulphidebondsarealsoresponsibleforthestability) e. Thesidechainarefreetorotateoneithersideofthepeptidebond. f. Distancebetweenaminoacidsis1.32Awhichismidwaybetweenthatofsinglebond(1.49A)and doublebond(1.27A) g. Ramachandranangles:areangleofrotation→determinethespatialorientationofpeptidechain
- 12. Characteristics of Primary structure of proteins:2 5. In polypeptide chain , at one end there will be one free alpha amino group→ N- terminal end and protein biosynthesis starts from amino terminal end (the first amino acid ). 6. The other end of polypeptide chain , is carboxy terminal end (the last amino acid) where there is free alpha carboxy group. 7. All other alpha-amino acids and alpha-carboxy groups are involved in peptide bond formation. 8. Writing of peptide structures –by convention ,the amino acid sequence is written from left to right with the free amino end (N- terminal acid/residue→ number 1 by tradition) on left and ending with the free carboxyl end (C –terminal amino acid / residue). 9. Shorthand to read peptides : three letter or one letter abbreviation/ short hand form of amino acids in protein to be read from N-terminal residue on left of peptide e.g. (Gly-Ala–Val)→Glycyl –Alanyl –Valine → G A V Or NH2-Gly-Ala-Val-COOH 10. Naming peptides : for naming peptides the amino acid Suffixes ine (Glycine → to glycyl) , an (Tryptophan), ate ( Glutamate→ Glutamyl) by yl with the exception of C- terminal amino acid.
- 13. Use of symbols in representing a peptide • A tripeptide → 3 amino acids and 2 peptide bonds is shown. H3 N + Glutamate – Cysteine –Glycine -COO- E – C – G Glu – Cys – Gly Glutamyl – cysteinyl –Glycine • Free amino end (N- terminal acid/residue) →-N + H3 is on the left. Free carboxyl end (C –terminal amino acid / residue)→ -COO- is on right . • The amino acid sequence is written and read from left to right. This is the chemical shorthand to write proteins. one letter abbreviation three letter abbreviation Peptide name Amino acids in a peptide
- 14. Polypeptide chain showing N-terminal and C-terminal • Schematic diagram NH3 + – CH – CO –NH –CH – CO –NH –CH – CO –NH –CH – CO –NH- CH – COO- I I I I I R1 R2 R3 R4 R5 N- terminal peptide bond amino acid residue C - terminal N C
- 15. Amino acid Amino acid composition ofHuman Cytochrome C (104AA) Amino acid compositionof Bovine Chymotrypsinogen (245AA) Ala 6 22 Arg 2 4 Asn 5 15 Asp 3 8 Cys 2 10 Glu 8 5 Gln 2 10 Gly 13 23 His 3 2 Ile 8 10 Amino acid Amino acid compositionof Human Cytochrome C (104AA) Amino acid compositionof Bovine chymotrypsinogen (245AA) Leu 6 19 Lys 18 14 Met 3 2 Phe 3 6 Pro 4 9 Ser 2 28 Thr 7 23 Trp 1 8 Tyr 5 4 Val 3 23
- 16. Clinical applications of primary structure ❖Clinical applications of primary structure : 1. Presence of specific amino acid at a specific position/number is very significant for a particular function of protein . Any change in the sequence is abnormal and may affect the functions and properties of proteins . 2. Many genetic diseases result from protein with abnormal amino acid sequences. If the primary structure of the normal and mutated proteins are known ,the this information may be used to diagnose or clinical study of the disease.
- 17. Secondary structure of proteins 3 dimensional Conformation of the polypeptide chain by twisting or folding is referred to as a secondary structure. (Types :Alpha-helix ,Beta-pleated sheet)
- 18. Secondary Structures of proteins • Secondary Structures :spatial arrangement of proteins by twisting of polypeptide chain= folding /helical coiling patterns in proteins (alpha- helix) or zig-zag linear( beta-sheet) or mixed form by hydrogen bonding and disulphide bonds . • Secondary Structures denotes the steric relationship of amino acids close to each other. • One of the form of coiling of the polypeptide chain is right handed alpha- helix. • Since proteins are made up of L-amino acids , the coiling of polypeptide chain into right handed alpha-helix is facilitated. • Super secondary Structures :indicate folding patterns in proteins • Linus Pauling (Noble 1954)and Robert Corey (Noble 1951) proposed alpha- helix and beta-pleated sheet structures of polypeptide chains.
- 19. Primary and Secondary structure of Human Insulin Carboxy-terminalend A chain : Asparagine B chain : Threonine Amino-terminal end A chain : Glycine B chain : Phenylalanine A polypeptide chain : 21 amino acids B polypeptide chain : 30 amino acids intrachain disulphide bond : between Cysteine residues( 6 th amino acid of A chain with 11 th amino acid of B chain) interchain disulphide bonds
- 20. Different kinds of secondary structures ❖Different kinds of secondary structures: 1. Alpha-helix (helicoid state) 2. Beta-pleated sheet (stretched state) 3. Loop regions 4. Beta-bends or beta-turns 5. Disordered regions 6. Triple helix
- 21. Alpha()-helix • Alpha()-helix : is called Alpha() because the first structure elucidated by Linus Pauling (Noble 1954)and Robert Corey (Noble 1951).It is most common spatial structure of protein . • If a backbone of polypeptide chain is twisted by equal amounts about each -carbon , it forms a coil or helix . The -helix is a rod like structure . • These hydrogen bonds have an essentially optimal nitrogen to oxygen (N-O) distance of 2.8 A. Thus, carbonyl(CO)group of each amino acid is hydrogen bonded to the -NH of the amino acid that is situated 4 residues ahead in a linear sequence . • The axial distance between adjacent amino acids is 1.5 A and gives 3.6 amino acid residues per turn of helix.
- 22. Characteristics of Alpha-helix (a type of Secondary structure of proteins):1 ❖Characteristics of Alpha-helix : 1. The most common stable conformation formed spontaneously with the lowest energy. 2. right or left handed Spiral/helical /tightly coiled structure in the stable form. A right handed helix turns in the direction that the fingers of right hand curl when its thumb points in the direction the helix rises. 3. Stabilized by Hydrogen bonds (weak, strong enough due to large number to stabilize alpha helix structure) of the main chain which forms the back bone. Side chains of amino acids extend outwards from the central axis. 4. Hydrogen bonding occurs between the carboxyl oxygen of one peptide bond and the amide nitrogen of another peptide bond and 3 amino acid residues apart / further down in the chain (e.g. 5th is hydrogen bonded to 9th and 6th is bonded 10th and so on). All peptide bonds except the first and the last in polypeptide chain participate in hydrogen bonding. 5. Each peptide bond in the polypeptide chain participates in intrachain hydrogen bonding.
- 23. Characteristics of Alpha-helix (a type of Secondary structure of proteins):2 6. Each turn of Alpha-helix : a. contains 3.6 amino acids residues /turn of the helix with the R group protruding outward radially. b. A rise along the central axis of 1.5A per residue and travels distance of 5.4 nm/turn. c. Spacing of each amino acid residue ( axial distance between amino acids) is 0.15nm (1.5 A translation). 6. The right handed -helix more stable and common than left handed helix. Left handed helix are rare because of presence of L-amino acid found in protein which exclude left handedness). 7. Proline , hydroxy proline and Glycine disrupt alpha-helix formation and introduce a kink or a bend in the helix. 8. Large number of acidic (Asp,Glu) or basic amino acids (Lys , Arg ,His)or amino acid with bulky R group disrupt Alpha-helix. 9. Abundant in Myoglobin , Hemoglobin , Keratin of hair (-Keratin ), proteins in wool and virtually, absent in Chymotrypsin.
- 24. Right and left-handed alpha-helix structure in protein molecule • A Right-handed alpha-helix turns in direction that fingers of right hand curl when thumb points in direction of helix rises. • A left- handed alpha-helix turns in direction that fingers of left hand curl when thumb points in direction of helix rises.
- 25. Schematic diagram of alpha-helical structure of proteins Alpha-helix :EachoxygenofC=Ogroupofapeptidebondformsahydrogenbondwiththehydrogenatom attachedtothenitrogeninapeptidebond,fouraminoacidsfurtheralongthepolypeptidechain. Each turn of Alpha- helix travels distance of 5.4 nm/turn. Alpha-helix
- 26. Alpha-helix (a type of Secondary structure of proteins):3 Alpha-helix:Hydrogenbondingoccursbetweenthecarboxyloxygenofonepeptidebondandtheamide nitrogenof another peptidebondand3aminoacidresiduesapart/furtherdowninthechain(e.g. 5th is hydrogenbondedto9th and6th isbonded10th andsoon).Allpeptidebondsexceptthefirstandthelastin polypeptidechainparticipateinhydrogenbonding.
- 27. Schematic diagram of alpha-helical structure of proteins Alpha-helix:StabilizedbyHydrogenbonds(weak,strongenoughduetolargenumbertostabilizealpha-helix structure)ofthemainchainwhichformsthebackbone.Sidechainsofaminoacidsextendoutwardsfrom thecentralaxis. Sidechainsofaminoacidsextend outwardsfromthecentralaxis. StabilizedbyHydrogen bonds
- 28. Examples of proteins with alpha-helical structure ❖Alpha-helical structure occur in both fibrous and globular proteins. Fibrous proteins with alpha-helical structure Globular proteins with alpha-helical structure -Keratin of hair , nail , skin Hemoglobin (80%) Fibrin of blood Myoglobin Myosin and Tropomyosin of muscle Proteins in wool Digestive enzyme→ Chymotrypsin is virtually devoid of alpha- helix in its structure.
- 29. Formation of hydrogen bond in alpha()-helix NH3 + – CH – CO –NH –CH – CO –NH –CH – CO –NH –CH – CO –NH- CH – COO- I I I I I R1 R2 R3 R4 R5 ---------Hydrogen bond ( N-O distance =2.8 A)------------- 3.6 amino acid residues
- 30. Helix destabilizing amino acids ❖Helix destabilizing (helix beakers)amino acids : Glycine , Proline ❖Proline as a helix beaker : since nitrogen of Proline residue in a peptide linkage has no substituted hydrogen (as it has imino NH-group instead of amino group) for the formation of hydrogen bond with other residue , Proline fits only the first turn of an alpha-helix. Elsewhere, it produces bend and turn. ❖Glycine as a helix beaker : all bends in alpha-helix are not caused by Proline residue but bend often occurs also at Glycine residues as side chain of Glycine is small.
- 31. Peptide bond formation with Proline Schematic diagram H H NH2 – C – CO N C – COOH → NH2 –CH – CO – C – COOH + I I R1 R1 Amino acid Proline peptide nitrogen of Proline has no substituted hydrogen
- 32. Structural importance of Alpha-helix ❖Structural importance of alpha-helix : Several alpha-helices can coil around one another like a twisted twined cable forming strong stiff bundles of fibers and give mechanical support.
- 33. Characteristics of Beta-pleated sheet (Secondary structure of proteins) ❖Characteristics of Beta-pleated sheet :where proposed/described by Linus Pauling (Noble 1954)and Robert Corey (Noble 1951). It is Beta because , it was the second type of structure after alpha-helix they elucidated. 1. When alpha-helix of keratin is stretched the hydrogen bonds are broken and new hydrogen bonds are formed between -CO and NH- of adjacent parallel chain / neighboring polypeptide segments giving rise to an arrangement of the backbone of protein molecule→ Beta-pleated sheet. (Beta-sheet appear pleated). 2. is stabilized by the hydrogen bonds . 3. Hydrogen bonding occurs between two polypeptide chains(H-bonds are intrachain) or two regions (neighboring segments) of a single chain of polypeptide chain (H-bonds are interchain). 4. Composed of 2 or more segments of fully extended polypeptide chains. 5. Two polypeptide chains in a beta-pleated sheet may run in the same direction (parallel beta-pleated sheets ) with regard to amino and carboxy terminal ends of polypeptide chain or in the opposite directions (anti-parallel beta-pleated sheets). 6. Distance between adjacent amino acid residue is 3.5 A(translation). 7. Major structural motif in fibroin of silk (anti-parallel), flavodoxin (parallel), Carbonic anhydrase(both) ,some regions of globular proteins like chymotrypsin, ribonuclease.
- 34. Intrachain hydrogen bonds (within single polypeptide chain) and interchain hydrogen bonds (between two or more polypeptide chains ) Interchainhydrogen bonds:areformedbetweenamidehydrogen( NH)ofonepolypeptidechain andcarbonyl(C=O)ofneighboringpolypeptidechain. Intrachainhydrogen bonds:areformedbetweenamidehydrogen( NH)ofonepolypeptidechain andcarbonyl(C=O)ofthesamepolypeptidechain.
- 35. ComparisonofAlpha-helix and Beta-pleated sheet: secondary structure of proteins Criteria Alpha-helix Beta-pleated sheet Structure Coiledrodlike Fullyextendedsheetlike Axialdistancebetweenaminoacids 1.5A 3.5A Numberofconstituentpolypeptidechain One Oneormorepolypeptide chains Stabilizedbyhydrogenbondsbetween NHandC=Ogroupsin thesame polypeptidechain Different/thesame polypeptidechain hydrogenbondsare Parallelto polypeptide backbone Perpendicularto polypeptidebackbone
- 36. ComparisonofAlpha-helixandBeta-pleatedsheet:secondarystructuresofproteins hydrogenbondsarePerpendicularto polypeptidebackbone. hydrogenbondsareParallel topolypeptidebackbone Coiledrodlike Fullyextendedsheetlike Axialdistancebetween aminoacids 1.5A Axialdistancebetweenaminoacids3.5A Oneconstituentpolypeptide chain Alpha-helix Beta-pleated sheet Oneormoreconstituentpolypeptidechain
- 37. Alpha-helix ,Beta-pleated sheet :secondary structures of important proteins Alpha-helix : abundant in Myoglobin , hemoglobin , Keratin of hair (- Keratin), proteins in wool and absent in chymotrypsin Beta-pleated sheet : fibroin of silk ,some regions of globular proteins like chymotrypsin, ribonuclease
- 38. Thearrangementofpolypeptidechainsinbeta-pleatedsheetconformation ❖The arrangement of polypeptide chains in beta- pleated sheet conformation can occur two ways: 1. Parallel beta- pleated sheet 2. Anti-parallel beta- pleated sheet ➢A beta- sheet can also be formed by either a single polypeptide chain folding back on to itself (H-bonds are intrachain and stabilized by intramolecular hydrogen bonding ) or separate polypeptide chains (H-bonds are interchain) . ➢As such , the -helix and -sheet are commonly found in the same protein structure . In the globular proteins , -sheet form the core structure.
- 39. Intrachain hydrogen bonds (within single polypeptide chain) and interchain hydrogen bonds (between two or more polypeptide chains ) Interchainhydrogen bonds:areformedbetweenamidehydrogen( NH)ofonepolypeptidechain andcarbonyl(C=O)ofneighboringpolypeptidechain Intrachainhydrogen bonds:areformedbetweenamidehydrogen( NH)ofonepolypeptidechain andcarbonyl(C=O)ofthesamepolypeptidechain
- 40. Hydrogen bonds in beta-pleated sheet structure H O H N C N C N C O H O C N C O I II I II II II II I I • • • • H O • •N HI N I H • C Hydrogen bond between chains Beta-pleated sheets(or simply sheets )are composed of two or more segments of fully extended peptide chains. Schematic diagram
- 41. Parallel and anti-parallel beta- pleated sheet Parallel beta- pleated sheet :same direction of N & C- terminal ends of peptide/Two polypeptide chains run in the same directions (parallel). Anti-parallel beta-pleated sheet: Opposite direction of N & C- terminal ends peptide/ Two polypeptide chains run in the opposite directions (antiparallel).
- 42. Structure of beta-pleated sheet N-terminal C–terminal N-terminal C-terminal C-terminal N-terminal The polypeptide chains in the beta() –sheets may be arranged either in parallel (the same direction) or anti-parallel (opposite direction). Parallel beta- pleated sheets Antiparallel beta- pleated sheets
- 43. Properties of Beta-pleated sheet (Secondary structure of proteins) Parallel beta- pleated sheet: the polypeptide are side by side and lie in same direction of N & C-terminal ends of peptide, so that their terminal residues are at the same end ( N-terminal faces to N-terminal ). It is stabilized by intrachain hydrogen bonds. Anti-parallel beta- pleated sheet : Opposite direction of N & C-terminal ends peptide.
- 44. Anti-parallel beta-pleated sheet: the polypeptide lie in opposite directions i.e. N – terminal end of one polypeptide is next to C – terminal of the other. (N- terminal faces C-terminal end of peptide- Anti- parallel directions ). It is stabilized by interchain hydrogen bonds. Anti parallel pleated beta-sheet of Secondary structure of proteins
- 45. Clinical application of beta-pleated sheet : Secondary structure of proteins ❖Clinical application of beta- pleated sheet : Secondary structure of proteins found in both fibrous and globular proteins. • Anti-parallel beta-sheets conformation is less common in human proteins. ❖Occurrence of beta-sheet : 1. Silk fibroin(the best example in nature) 2. Amyloid in human tissue: a protein that accumulates in Amyloidosis and Alzheimer’s disease .Dementia occurring in middle age associated with this Amyloidosis .
- 46. Loop regions and their Importance ❖Loop regions : • About half of the residues in a typical globular protein are present in alpha- helices or beta-sheet . Remaining residues are present in loop or coil conformation. Loop regions though irregularly ordered (lacking regular secondary structure) are biologically important as they are more ordered secondary structure. • Loop or coiled the random coils (disordered and biological unimportant conformation of denatured proteins). ❖Importance of loop regions : form the antigen-binding sites of antibodies.
- 47. Beta-bend or beta-turn and its Importance ❖Beta-bend or beta-turn or hairpin turn or reverse turn : refers to the segment , in which a polypeptide chain abruptly reverses direction and often connects the ends of the adjacent antiparallel beta-strands hence they are named as beta-turn. Globular proteins contain Beta-bends. ❖Characteristics of beta-bend(-bend) : 1. consists of four successive amino acid residues. 2. Frequently contains Proline or Glycine or both. 3. is stabilized by Intrachain disulphide bridges and hydrogen bonds (hydrogen bond is formed between the first amino acid to the forth in the bend). 4. occur primarily at protein surfaces and impart globular shape (rather than linearity) to proteins. 5. Promote the formation of anti-parallel beta –pleated sheets . 6. Importance of beta-bends : they help in the formation of compact globular structure.
- 48. Beta bends & non –repetitive secondary structure of proteins Beta-bends may be formed in many proteins by the abrupt U-turn folding of the chain. Intrachain disulphide bridges and hydrogen bonds stabilize these bends .
- 49. Disordered region and its Importance • Not all residues are necessarily present or ordered secondary structure. • Specific residues of many proteins exist in numerous conformation in solution and thus they are called Disordered regions. • Many Disordered region become ordered region when a specific ligand is bound . • e.g. The stabilization of Disordered regions of the catalytic sites of many enzymes when ligand is bound . • Importance of Disordered region : gives flexibility and performs a vital biological role.
- 50. Disordered region of enzyme and its Importance Schematic diagram Many Disordered region become ordered region when a specific ligand is bound . Disordered region gives flexibility and performs a vital biological role. Disordered regions of enzyme- catalytic site ordered regions of enzyme Substrate added to enzyme catalyzed reaction mixture Enzyme with Disordered regions set free at the end of reaction Product Enzyme-substrate complex –induced fit +
- 51. Super secondary structures of proteins ❖A protein molecule may contain all types of arrangements in different parts . Thus , a part may form an -helix to be followed by -pleated sheets which may include parallel or anti- parallel regions with intervening turns ,loop regions and disordered regions. Such combinations of secondary structural features are called Super secondary structures. ➢These grouping of certain secondary structural elements of proteins occur in many unrelated globular proteins.
- 52. Characteristic properties of Super secondary structures of proteins ❖Characteristic properties of Super secondary structures of proteins: 1. Folding patterns involving - helices, -pleated sheets (which may be parallel or anti- parallel regions with intervening turns) ,loop regions and disordered regions. 2. - - 2 : in this structure ,an -helix connects two parallel strands of -pleated sheets. It is the most common motif. 3. -hairpin : consists of antiparallel -sheets joined by relatively tight reverse turn/ short loops. 4. motif: two successive anti-parallel helices packed against each other with their axis inclined. 5. -barrel: extended -pleated sheets role up to form three different types of barrels 6. Globular proteins like Chymotrypsin , Myoglobin and Ribonuclease have Super secondary structures instead of uniform secondary structures. 7. The secondary and Super secondary structures of large proteins are recognized as domains or motifs.
- 53. Domains of the globular proteins ❖Domains of the globular protein : ➢The term domain is used to represent the basic structural and functional units of protein with tertiary structure(denotes a compact globular functional unit of protein). ➢Relatively independent region and may represent a functional unit. ➢ are usually connected with relatively flexible areas of protein (e.g. immunoglobulin) ❖3 Domains of Phenylalanine hydroxylase : a. Catalytic b. Regulatory c. Protein-protein interaction domain ❖Calmodulin : a calcium binding regulatory protein(regulates intracellular calcium levels ) ❖A polypeptide with 200 amino acids normally consists of two or more domains.
- 54. - - 2 Protein motifs - - 2 : an -helix connects two parallel strands of -pleated sheets. Folding patterns involving - helices and -pleated sheets.
- 55. motif and -hairpin Protein motifs(Super secondary structure of proteins) motif: two successive anti-parallel helices packed against each other with their axis inclined. -hairpin : consists of antiparallel -sheets joined by relatively tight reverse turn/ short loops.
- 56. -barrel Protein motifs(Super secondary structure of proteins) -barrel: extended -pleated sheets role up to form different types of barrels Role of Super secondary structure of membrane proteins
- 57. -barrels located across Mitochondrial outer membrane -barrels located across the Mitochondrial outer membrane facilitate transport of moieties associated with functions of mitochondrion (e.g. Biological oxidation , Urea cycle etc. ).
- 58. Protein motifs(Super secondary structure of proteins)
- 59. Predominant Specific structural motifs of common proteins Protein Predominant Specific structural motifs present Myoglobin Alpha-helix and beta-pleated sheets Flavodoxin Parallel beta-pleated sheets Super oxide dismutase Anti-parallel beta-pleated sheets Fibroin of silk anti-parallel beta-pleated sheets Carbonic anhydrase Both Parallel and Anti-parallel beta-pleated sheets Keratin Alpha-helix →Coiled coil Collagen Triple-helix Elastin No specific motif
- 60. Approximateamountofaalpha-helixandbeta-structureinsome singlechainprotein Protein Total residue Alpha-helix (residue%) Beta-structure (residue%) Myoglobin 153 78 0 Cytochrome C 104 39 0 Lysozyme 129 40 12 Ribonuclease 124 26 35 Chymotrypsin 247 14 45 Carboxy peptidase 307 38 17 Data from C. R. Cantor and P.R. Schimnel ,Biophysical chemistry , the conformation of biological macromolecules p 100 1980
- 61. Tertiary Structure of proteins Tertiary Structure : Three dimensional structure generated by interaction between the amino acid residues of functional proteins.
- 62. Tertiary Structure of proteins ❖Tertiary structure of globular proteins defines the steric relationship of amino acids which are far apart from each other in the linear sequence but are close in three dimensional aspects i.e. Three-dimensional structure of globular proteins is dependent on the primary structure. • It is a compact structure with hydrophobic side chains held interior while hydrophilic groups are on the surface of the protein molecule. This arrangement ensures stability of the molecules. • Three-dimensional structural conformation of globular proteins provides and maintains the functional characteristics. • Functions of globular proteins are maintained because of their ability to recognize and interact with a variety of molecules . • This structure reflects the overall shape of the molecule. • Primarystructureofproteinisfoldedtoformcompact,biologicallystableandactive conformation i.e. a three-dimensional globularprotein. It is referred as its Tertiary structure. • e.g. Insulin ,Myoglobin
- 63. Tertiary structure of Human Insulin In1953,FrederickSanger determinedprimarystructureofInsulin(apancreaticproteinhormone)and showed forthefirsttimethataproteinhasapreciselydefinedaminoacidsequence(primarystructure). PrimarystructureofHumanInsulinisfoldedtoformcompact,biologicallystableandactiveconformation i.e. a three-dimensionalglobularprotein.It is referred as its Tertiary structure.
- 64. Tertiary Structure of Myoglobin(Mb) Myoglobin:PrimarystructuresimilartosinglemonomericunitofHemoglobin withasinglepolypeptidechainhaving 153aminoacids(molecularweight16700).It haseightalpha–helices(AtoH)andonehemegroup(ironcontaining porphyrin)tofacilitateitsfunctionofoxygenstorageincardiacandskeletalmusclesinhumanbody,WhalesandSeals.
- 65. Tertiary Structure of Hemoglobin (Hb) Hemoglobin:Tetramericwith4hemegroups.Eachpolypeptidechainhassimilarstructureto single polypeptidechainofMyoglobin.Ithasaloweraffinity foroxygenthanMyoglobin.Foursubunits of Hb functioncooperatively.TetramericstructureofhemoglobinfacilitatessaturationwithO2inthelungand releaseofoxygenasittravelsthroughthecapillarybed.
- 66. Covalentbondsstabilizing proteinstructure ❖Proteins are stabilized by three types of covalent bonds: 1. Peptide bonds 2. Disulphide bonds 3. Lysinonorleucine bonds
- 67. Peptide bonds : Covalentbondsstabilizing proteinstructure • Formation of peptide bond = a covalent bond is formed by amide linkage between the - carboxyl group of one amino acid and - amino group of another amino acid by removal of a water molecule. • Successive amino acids are joined/cemented by peptide bonds ( -CO-NH) in proteins . ➢The peptide bonds form strong backbone of polypeptide and side chains of amino acid residue project outside the peptide backbone. Dipeptide :two amino acids and one peptide bond ( not two bonds)
- 68. Lysinonorleucine bonds: Covalent bonds responsible for protein structure ❖Lysinonorleucine bonds: • A bond formed between oxidized lysine residue with unmodified lysine side chain to form a crosslink, both within and between the triple helix units. e.g. Collagen • Cross links confer tensile strength to the protein .
- 69. Structure of collagen Type 1 ❖ Structure of collagen Type 1: 1. Triple-stranded helical structure present throughout the collagen molecule 2. Shape : rod-like molecule → 1.4 nm diameter and 300 nm length 3. Number of Amino acid residues : 1000 per for each polypeptide chain (3000 /molecule) 4. Amino acid contribution : 1/3 rd of amino acids are Glycine (every third amino acid in collagen is Glycine. 5. Repetitive amino acid sequence : (Gly – X –Y )n ,where X and Y represent other amino acids 6. Proline and hydroxyproline : 100 per for each polypeptide chain 7. Function of Proline and hydroxyproline : confer rigidity to the collagen molecule 8. Collagen Fibril formation : Triple helical molecule of collagen assemble to form elongated fibrils . It occurs by a quarter staggered alignment i.e. each triple helix is displaced longitudinally from its neighbor collagen molecule by about one-quarter of its length 9. Collagen Fiber formation : Collagen Fibrils assemble to form rod like fibers . 10. Strength of Collagen Fiber : contributed by covalent cross linking of formed between Lysine and hydroxylysine and also between Proline and hydroxyproline.
- 70. Disulfide bonds : Covalentbondsstabilizing proteinstructure ❖Proteins are stabilized by three types of covalent bonds (peptide bonds , disulphide bonds and Lysinonorleucine bonds). • Cysteine : with functional group -SH (sulfhydryl) • Cystine : functional group -S-S-(disulphide) • A covalent disulfide (-S-S)bond formed between the sulfhydryl group (-SH)of two Cysteine residues in the same or different polypeptide chains. • Intra chain S-S bond →Cysteine at 6th position linked to that at 11th position of A chain of Insulin ❖Inter chain S-S bonds : (1)Cysteine at 20th position of A chain is linked to that at 19th position of B chain of Insulin (2) Cysteine at 7th position of A chain is linked to that at 7th position of B chain of Insulin ➢These disulfide bonds contribute to structural conformation and stability of proteins . Performic acid cleaves the disulfide bonds between polypeptide units.
- 71. Role of Cysteine and Cystine in formation Covalentbondsstabilizing proteinstructure Cystine has disulfide (S-S) as a functional group . It is formed from Cystine (Dicysteine)after oxidation. Cystine on reduction yields two Cysteine molecules. Two Cysteine residues can connect two polypeptide chains by formation of Interchain disulfide bonds or links . e.g. Keratin , Insulin
- 72. Intrachain and Interchain Disulfide bonds (S-S bonds) in Human Insulin Carboxy-terminalend A chain : Asparagine B chain : Threonine Amino-terminal end A chain : Glycine B chain : Phenylalanine A polypeptide chain : 21 amino acids B polypeptide chain : 30 amino acids intrachain disulphide bond : between Cysteine residues( 6 th amino acid of A chain with 11 th amino acid of B chain) interchain disulphide bonds
- 73. ClassificationAlpha-keratinstructurebasedonSulphuranddisulphidebridgescontent Type of keratin abundant in structure Sulphur and disulphide bridges content Hard keratin Hair, nails ,horn High Sulphur and disulphide bridges (rich in cysteine residue) content Soft keratin Skin Low Sulphur and disulphide bridges (poor in cysteine residue) content ➢ Disulphide bonds are common in structural proteins like Keratin , extracellular enzymes such as ribonuclease but rare in Intracellular globular proteins. ➢ These bonds help to stabilize protein molecules against denaturation and confer additional stability them .
- 74. Importance of Disulphide bonds of Keratin structure in hair • Springiness of hair is due to the characteristic coiled coil structural motif. • When stretched , the coiled coil will untwist and resume the original structure. • Hair styling /dressing : Stretching of hair using moist heat to break disulphide bonds of keratin structure. • Epidermolysis bullosa : abnormalities in keratin structure →loss of integrity of skin Hair styling Hair dressing / status of disulphide bond of keratin Stretching hair styling Reduction of disulphide bonds of keratin to break these bonds Curling hair styling Re-oxidation of disulphide bonds of keratin to form new bonds
- 75. Hair styling /dressing : Stretching of hair using moist heat to break disulphide bonds of keratin structure Hard keratin and Soft keratin Stretching hair styling: Reduction of disulphide bonds of keratin to break these bonds High Sulphur and disulphide bridges (rich in cysteine residue) content
- 76. and -conformation of Keratin in Hair waving Hair with -helices of -Keratin -helices of -Keratin stretched → conformation of Keratin conformation of Keratin reverted to -helices of -Keratin cooling exposed to moist heat
- 77. Biochemical aspects of Hair curling(waving) Hair to be curled is bent to appropriate shape Disulfide bonds of Cystine are converted to sulfhydryl groups of Cysteine Uncoiling of -helical structure of -Keratin New Disulfide bonds are formed between Cysteine residues of keratin→ altered -helical structure of -Keratin Hair with desired curls (temporary structure) Growth of new hair with native conformation without curls Application of reducing agents Removal of reducing agents and addition of oxidizing agent Hair washed and cooled
- 78. Alteration of Keratin structure in hair waving I I I I I S S S S S I I I I I S S S S S I I I I I I I I I I SH SH SH SH SH SH SH SH SH SH I I I I I SH SH SH SH SH SH SH SH SH SH S S S S S S S SSH SH S S Reduction Curling Oxidation S S Schematic diagram
- 79. Hair styling /dressing Hair curling(waving) Hair straitening
- 80. Non-covalent interactions in Tertiary structure of globular proteins ❖ Tertiary structure of globular proteins : refers to the three-dimensional conformation of proteins generated and maintained by weak bonds(valence forces) / non-covalent interactions such as: a. Hydrogen bonds : formed between -CO and NH- of two different peptide bonds or -OH group of hydroxy amino acids(Serine etc.) and-COOH groups of acidic amino acids Aspartic or Glutamic acid. b. Ionic bonds/electrostatic interactions/salt bridges : formed between oppositely charged groups when they are in close vicinity . They are also formed oppositely charged R groups of polar amino acid residues . e.g. basic (Histidine, Arginine , Lysine)and acidic amino acids (Aspartic acid, Glutamic acid) c. Hydrophilic interactions : water loving groups are associated with water. d. Hydrophobic interactions : formed between hydrophobic groups (hydrocarbon like)of amino acids like Alanine and Phenylalanine. e. Van der Waal forces : weak ,but collectively contribute maximum towards the protein structure. ➢During folding of globular proteins (spherical/round), hydrophobic groups prefer to be interior and Hydrophilic groups prefer to be on the surface of protein molecule. ➢The tertiary structure acquired by a native protein is always thermodynamically most stable.
- 81. Properties of Hydrogen bonds responsible for protein structure ❖Properties of Hydrogen bonds responsible for protein structure: • Hydrogen bonds are formed between NH- and –CO groups of peptide bonds by sharing single hydrogen . • Each Hydrogen bond is weak but collectively they are strong. A large number of Hydrogen bonds significantly contribute stability to the protein structure. • Hydrogen bonds may be intrachain( between same polypeptide chain) or interchain( between different polypeptide chains) • Side chains of 11 out of 20 standard amino acids can participate in hydrogen bonding (i.e. Tryptophan, Tyrosine , Aspartic acid, Asparagine ,Glutamic acid, Glutamine , Histidine , Arginine , Serine , Threonine, Lysine). • Formation of Hydrogen bonds between polar side groups on the surface of protein enhances solubility of the protein.
- 82. Structure of interchain Hydrogen bonds H O H I II I Schematic diagram N C N C N C II I II O H O H H O C I N C II N C N O II I O H interchain Hydrogen bond III
- 83. Hydrogen donor and Hydrogen acceptor in Hydrogen bonds responsible for protein structure ❖Hydrogen bonds responsible for protein structure : are weak electrostatic interactions between one electronegative atom like O or N and hydrogen atom linked to second electronegative atom . Group of Hydrogen donor Hydrogen donor -NH Imidazole , indole ,peptide -OH Serine ,Threonine Group of Hydrogen acceptor Hydrogen acceptor COO- Aspartic acid ,Glutamic acid C=O Peptide -S-S- Disulphide
- 84. Hydrophobic bonds responsible for protein structure ❖Hydrophobic bonds in protein structure : ➢are formed by interactions between Hydrophobic R groups(non-polar side chains) of neutral amino acids like Alanine , Valine , Leucine, Isoleucine, Methionine, Phenylalanine , Tryptophan by eliminating water molecules. ➢are not true bonds. ➢ serves to hold lipophilic side chains together. ➢The occurrence of hydrophobic forces is observed in aqueous environment wherein the molecules are forced to stay together.
- 85. Hydrophobic interactions I I I NH-CH-CO HC-CH3 CH2 CH3 H3C CH3 CH CH2 NH-CH-CO I I Leucine Isoleucine Hydrophobic interactions: 1. non-polar side chains of neutral amino acids tend to be closely associated with each other in protein confirmation. 2. are Not true bonds. 3. Observed in aqueous environment wherein the molecules are forced to stay together. Schematic diagram
- 86. Electrostatic bonds: Bondsresponsibleforproteinstructure Electrostatic /ionic / salt bonds/salt bridges : formed between oppositely charged groups when they are in close vicinity i.e. negatively charged group linked to positively charged group of amino acid e.g. COO ⁻ of Glutamic acid associates with NH₃⁺of Lysine. They are also formed oppositely charged R groups of polar amino acid residues. Positive charges contributed by : epsilon amino group of lysine , guanidium group of Arginine, imidazole group of Histidine Negative charges contributed by : beta and gamma carboxyl group of Aspartic acid and Glutamic acid respectively
- 87. Electrostatic interactions NH-CH-CO CH C O O- N+ H3 (CH2)4 NH-CH2-CO Schematic diagram Aspartate: acidic amino acid Lysine: basicaminoacid ❖Electrostatic interactions: are formed between negatively charged groups ( e.g. COO -) of acidic amino acid with positively charged groups (e.g. -N+H3) of basic amino acid . ❖They are also formed oppositely charged R groups of polar amino acid residues. Electrostatic interactions → I I I I
- 88. Van Der Waals forces: Bondsresponsibleforproteinstructure ❖Van Der Waals forces/interactions : Electrically neutral molecules associate by electrostatic interactions due to induce dipoles. ❖Characteristics of Van Der Waals forces/interactions : • are the non-covalent associations. • are very weak ,but collectively contribute maximum towards the protein structure. • act only on short distances. • include both an attractive and repulsive component between both polar and non-polar side chain of amino acid residues.
- 89. Van Der Waals forces : Bondsresponsibleforproteinstructure Van Der Waals forces/interactions : Electrically neutral molecules associate by electrostatic interactions due to induce dipoles. These are the non-covalent associations. They are very weak ,but collectively contribute maximum towards the protein structure. They act only on short distances. They include both an attractive and repulsive component between both polar and non-polar side chain of amino acid residues. induce dipole
- 90. Factors stabilizing the tertiary structure of Globular proteins A B C D C=O S N O- H E F G H N + H3 S O Ionic interactions Covalent Cysteine interlinks Association of hydrophobic R groups within molecule, shielded from water Hydrogen bonds Schematic diagram
- 91. Weak bonds stabilizing Tertiary structure of proteins Peptide bond Serine Lysine Alanine Phenylalanine Cysteine NH CH2OH N+H3 CH3 S O O- O C=O C=O CH3 S C Peptide bond Asp Asp Alanine Phenylalanine Cysteine Hydrogen bonds ionic bond hydrophobic interactions disulfide bond = Tertiarystructureofanativeproteindenotesoverallarrangementandinter-relationshipofvariousregionsor domainsofsinglepolypeptidechain.Itisthermodynamicallythemoststableconformation. Schematic diagram
- 92. Domains of globular proteins ❖Domains of globular protein : ➢The term domain is used to represent the basic structural and functional units of protein with tertiary structure( denotes a compact globular functional unit of protein). ➢Relatively independent region and may represent a functional unit. ➢ are usually connected with relatively flexible areas of protein (e.g. immunoglobulin) ❖3 Domains of Phenylalanine hydroxylase : a. Catalytic b. Regulatory c. Protein-protein interaction domain ❖Calmodulin : a calcium binding regulatory protein(regulates intracellular calcium levels ) ❖A polypeptide with 200 amino acids normally consists of two or more domains.
- 93. Domains of globular proteinsimmunoglobulinsandPhenylalaninehydroxylase Domainsofimmunoglobulinsareusually connectedwithrelativelyflexibleareasofprotein. 3DomainsofPhenylalaninehydroxylase:Catalytic, Regulatory,Protein-proteininteractiondomain
- 94. Domains of Calmodulin : a calcium binding regulatory protein Domains of Calmodulin
- 95. Domains of ribonuclease Domains of ribonuclease :catalytic and RNA binding domain to facilitate its function of cleavage of ribonucleic acids(RNA)
- 96. Quaternary structure of globular proteins : refers to the spatial arrangement of subunits (polypeptide chains) linked by non- covalent interactions in protein molecule .
- 97. Quaternary structure of globular proteins:1 ❖A number of proteins are made up of with two or more peptide chains(subunits/ monomers / protomers )which may be identical or unrelated. Such proteins are termed as oligomers. Subunits of oligomers are not covalently linked(non-covalent forces). This association of subunits to form protein molecule is known as Quaternary structure. Not all proteins are polymeric. ✓Globular proteins loose their functions on dissociation of subunits. ❖Forces involved to aggregate subunits : a. Hydrogen bonds b. Ionic /electrostatic interactions c. Hydrophobic interactions d. Van der Waal forces ➢The same weak bonds are involved in secondary and tertiary structure in this associations.
- 98. Quaternary structure of globular proteins:2 ❖Quaternary structure of globular proteins :refers to the spatial arrangement of subunits (polypeptide chains) linked by non-covalent interactions in three dimensional complexes. It occurs in proteins with two or more peptide chains. . ❖Monomer or promoter or subunits :Individual polypeptide chain of oligomeric protein • Monomers in Quaternary structure of globular proteins stabilized by a. Hydrogen bonds b. Hydrophobic interactions non-covalent bonds c. Ionic bonds /electrostatic interactions/salt bridges d. Van der Waals forces ❖Dimer : 2 polypeptide chains (e.g. Insulin) ❖Tetramers : 4 polypeptide chains (e.g. LDH ,Hemoglobin ,SGOT) ❖Oligomers : proteins with 2 or more polypeptides chain
- 99. Examples of proteins(oligomers) having quaternary structure ❖Examples of proteins having quaternary structure : • Hemoglobin • Creatine kinase • Alkaline phosphatase • Glycolytic enzymes : a. Aldolase b. Lactate dehydrogenase c. Pyruvate dehydrogenase
- 100. Types of globular protein based on number of constituent polypeptide chains Type Component polypeptide chains as functional unit Exampleofglobularprotein Dimer 2 polypeptide chains Creatine kinase (CK) Homodimer contains 2 copies of the same polypeptide chain →2 Alpha() chains +2 Beta( )chains Hemoglobin Heterodimer contains 2 different types the polypeptides as a functional unit →with A and B polypeptide chains Insulin Tetramer 4 polypeptide chains ( H and M type polypeptide chains 2 each ) Lactate dehydrogenase (LDH) Tetramer 4 polypeptide chains ( light and heavy chains polypeptide chains 2 each ) Immunoglobulin G
- 101. Quaternary structure of Hemoglobin Hemoglobin (HbA1)has 4 Polypeptides chains (tetramer) associated by non-covalent bonds : 2 Alpha() chains + 2 Beta( )chains It possesses Quaternary structure(oligomeric). Inthis, Rgroupcontactsare presentbetweensimilarside chainsandthereisverylittle contactbetweendissimilar side chains. HbA: Tetramer 22 Eachchainhasoneheme groupandsooneFe2+ ion
- 102. Importance of Quaternary structure of globular proteins ❖Importance of Quaternary structure of globular proteins : 1. Subunits of oligomeric proteins may either function independently of each other or may work cooperatively as in Hemoglobin where the binding of oxygen to one subunit of tetramer increases the affinity of other subunits for oxygen. 2. Oligomeric proteins are regulators of cell metabolism & cellular functions.
- 103. Deoxy –Hb Hb O₂ Hb O₄ Hb O₆ Hb O₈ T form ↓ ↓ ↓ ↓ ↓ ↑ R form Oxygenation of Hemoglobin Quaternary structure of Hemoglobin favors its functions :Binding of oxygen(O2) to one heme unit facilitates oxygen binding by other subunits.
- 104. Four levels of structural organization of proteins Theoverallconformationofaprotein,theparticularpositionoftheaminoacidchainsinthreedimensional spacedeterminesthefunction/softheprotein.
- 105. Four levels of structural organization of proteins Many genetic diseases result from protein with abnormal amino acid sequences. If the primary structure of the normal and mutated proteins are known , this information may be used to diagnose or clinical study of the disease.