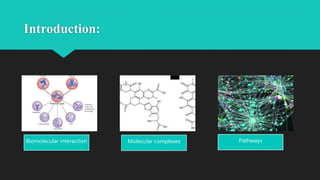
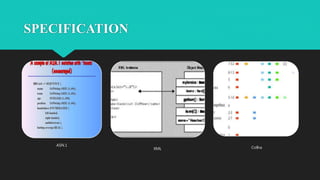
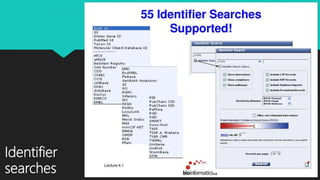
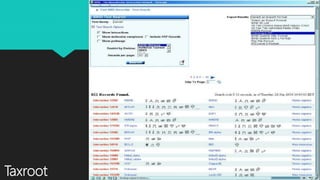
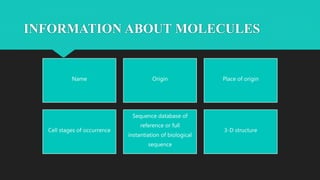
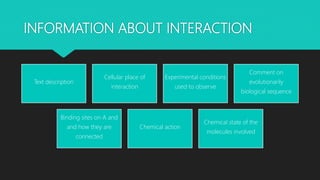
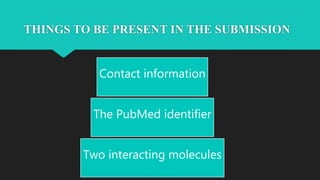
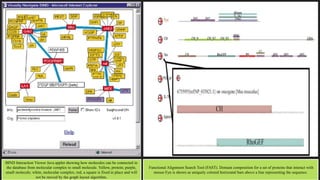
The document discusses the Biomolecular Interaction Network Database (BIND), which stores information about molecular interactions, complexes, and pathways. BIND uses standards like ASN.1 and XML to specify interactions. It stores details about molecules, interactions, publications, and more. Tools like Pajek and MCODE are used to visualize and analyze the network. The database has expanded to include additional details like post-translational modifications, cellular localization, and links to other databases. Manual and automatic submission is supported.