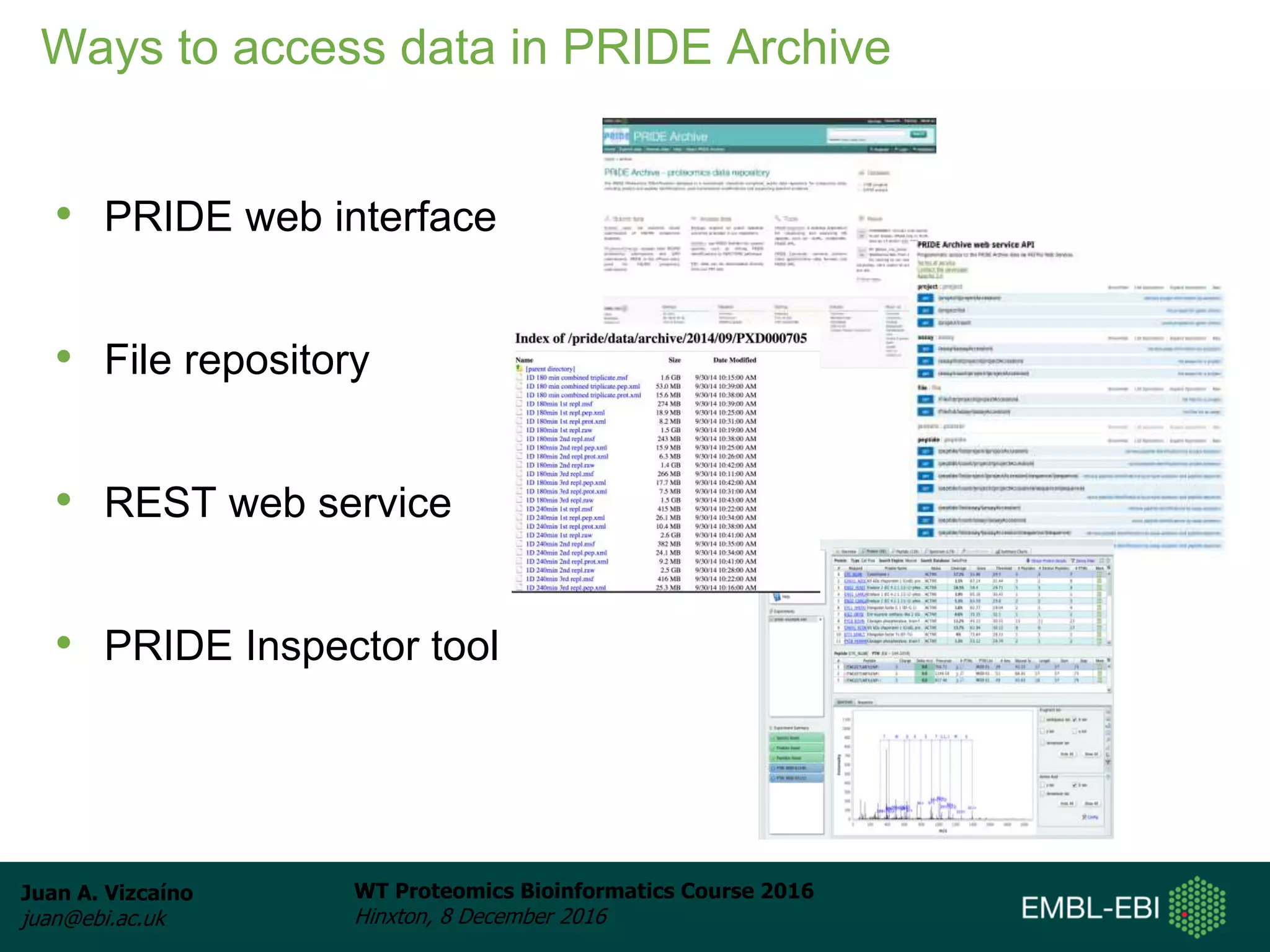
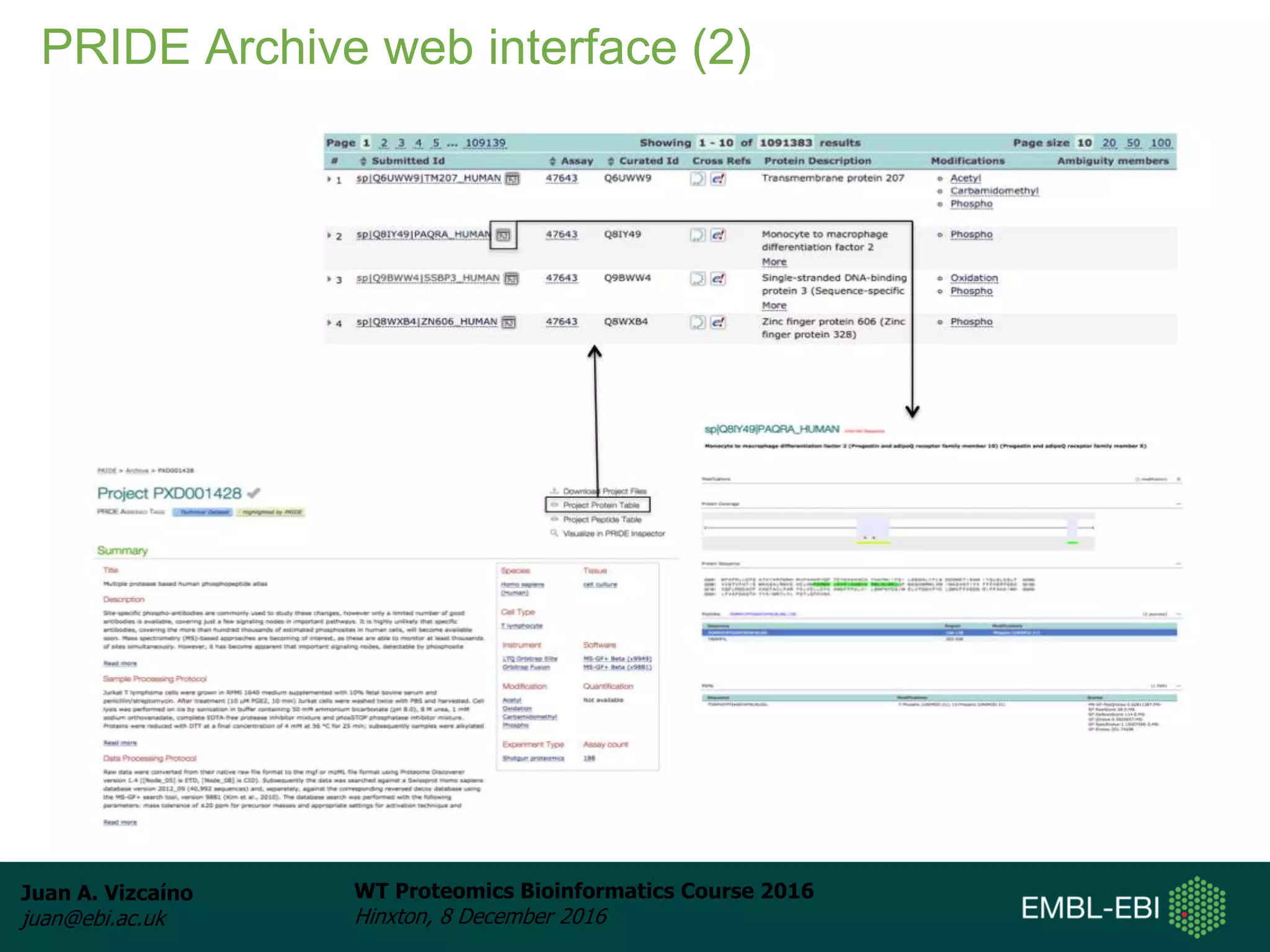
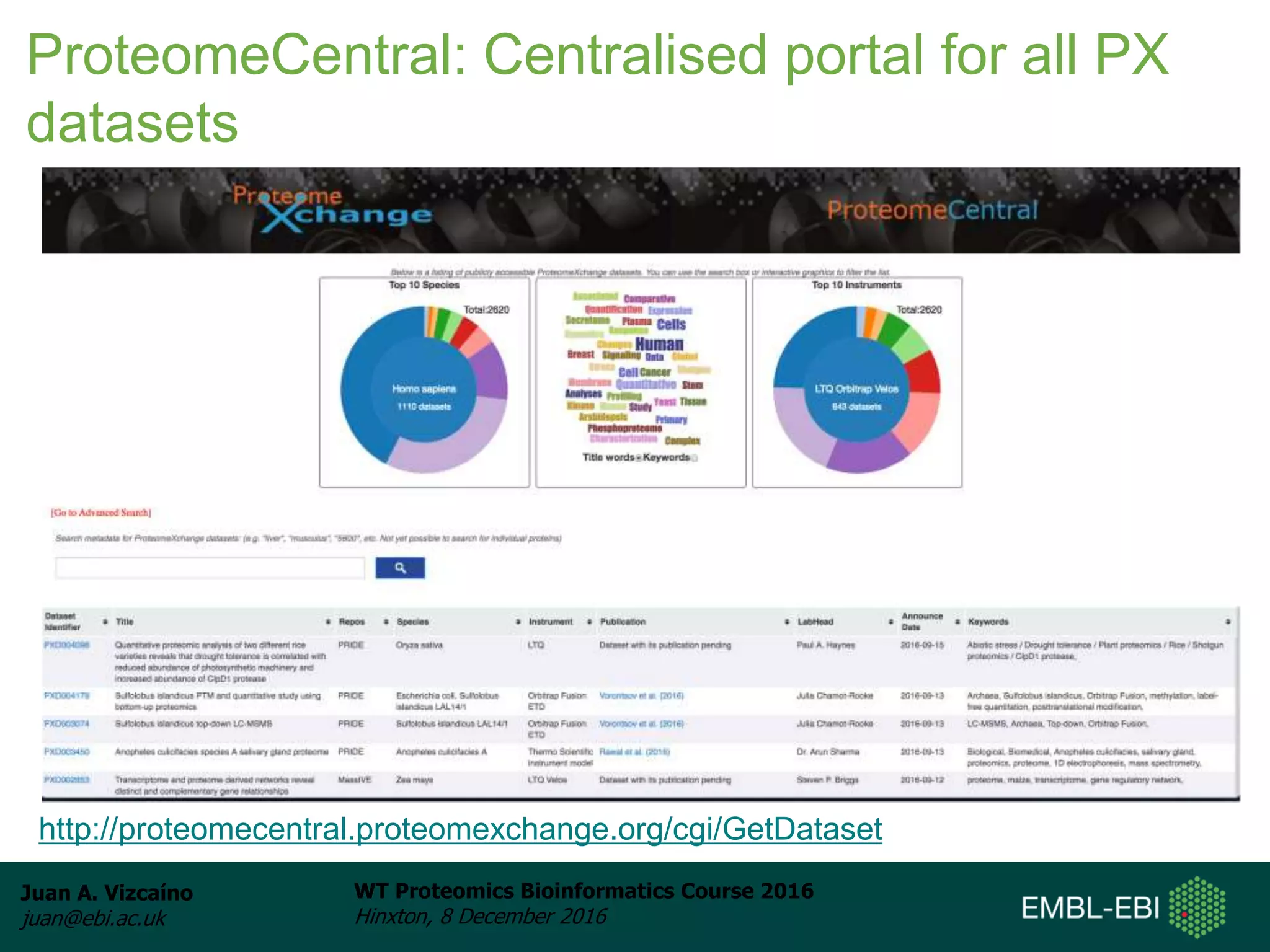
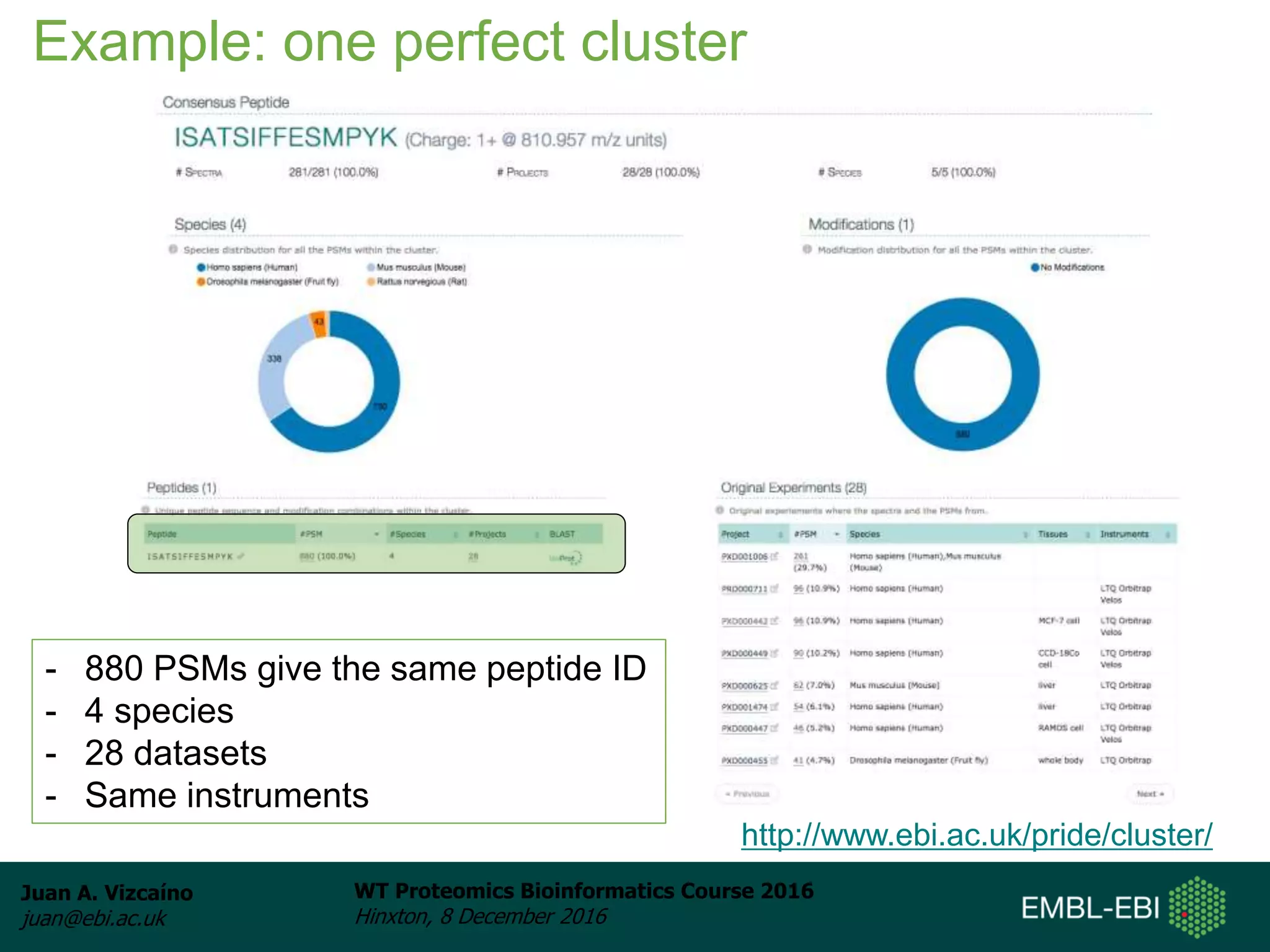
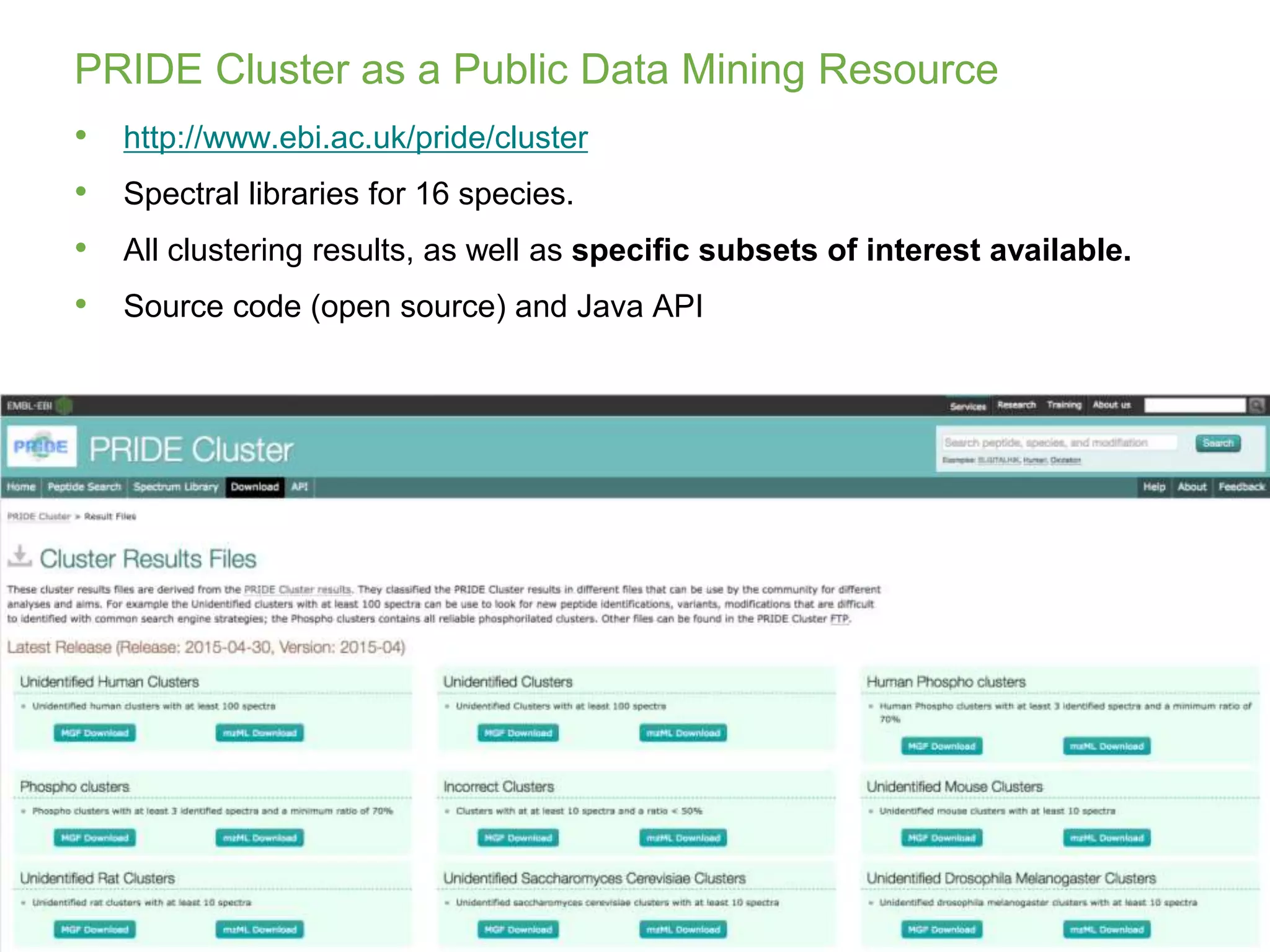
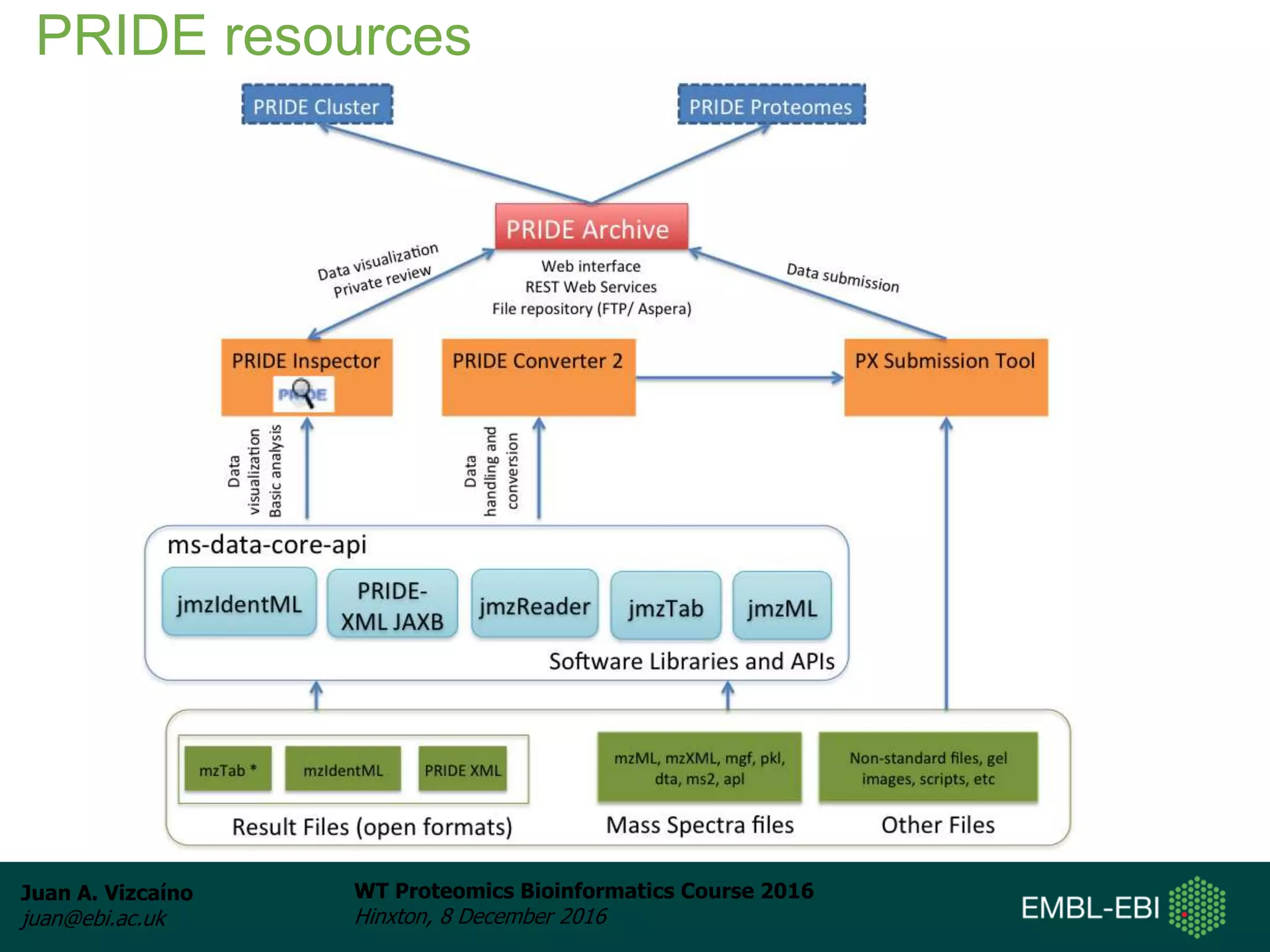
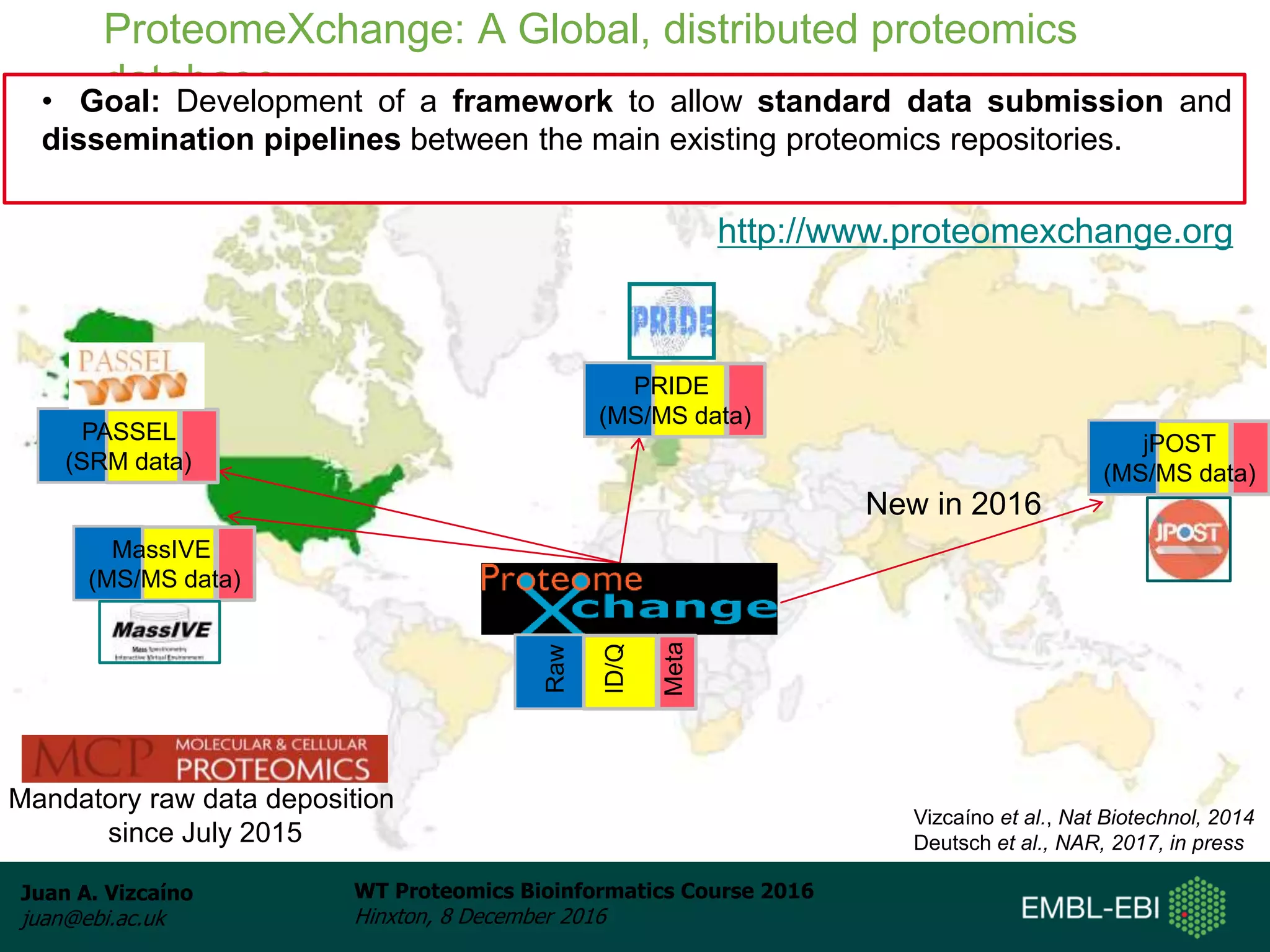
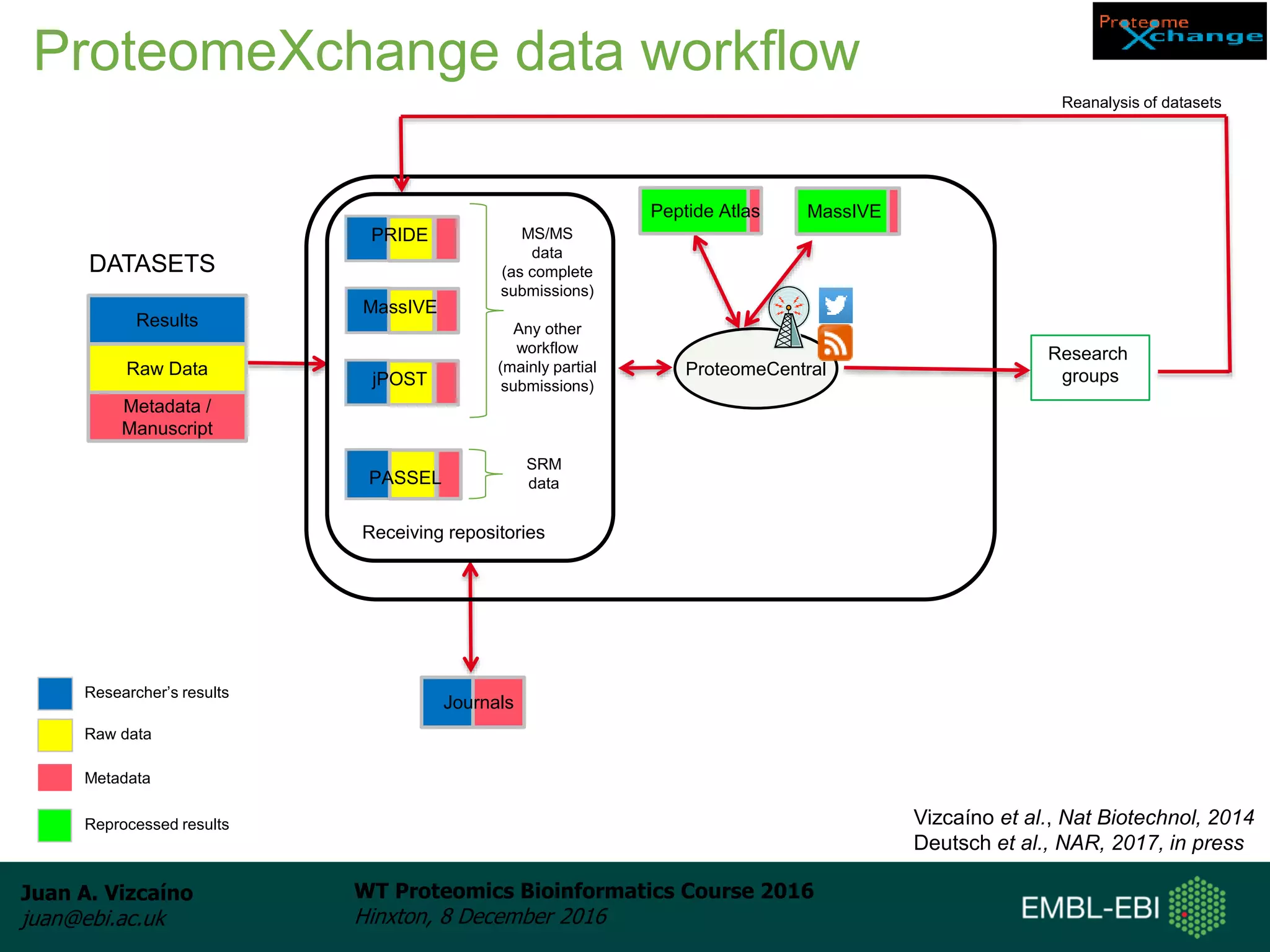
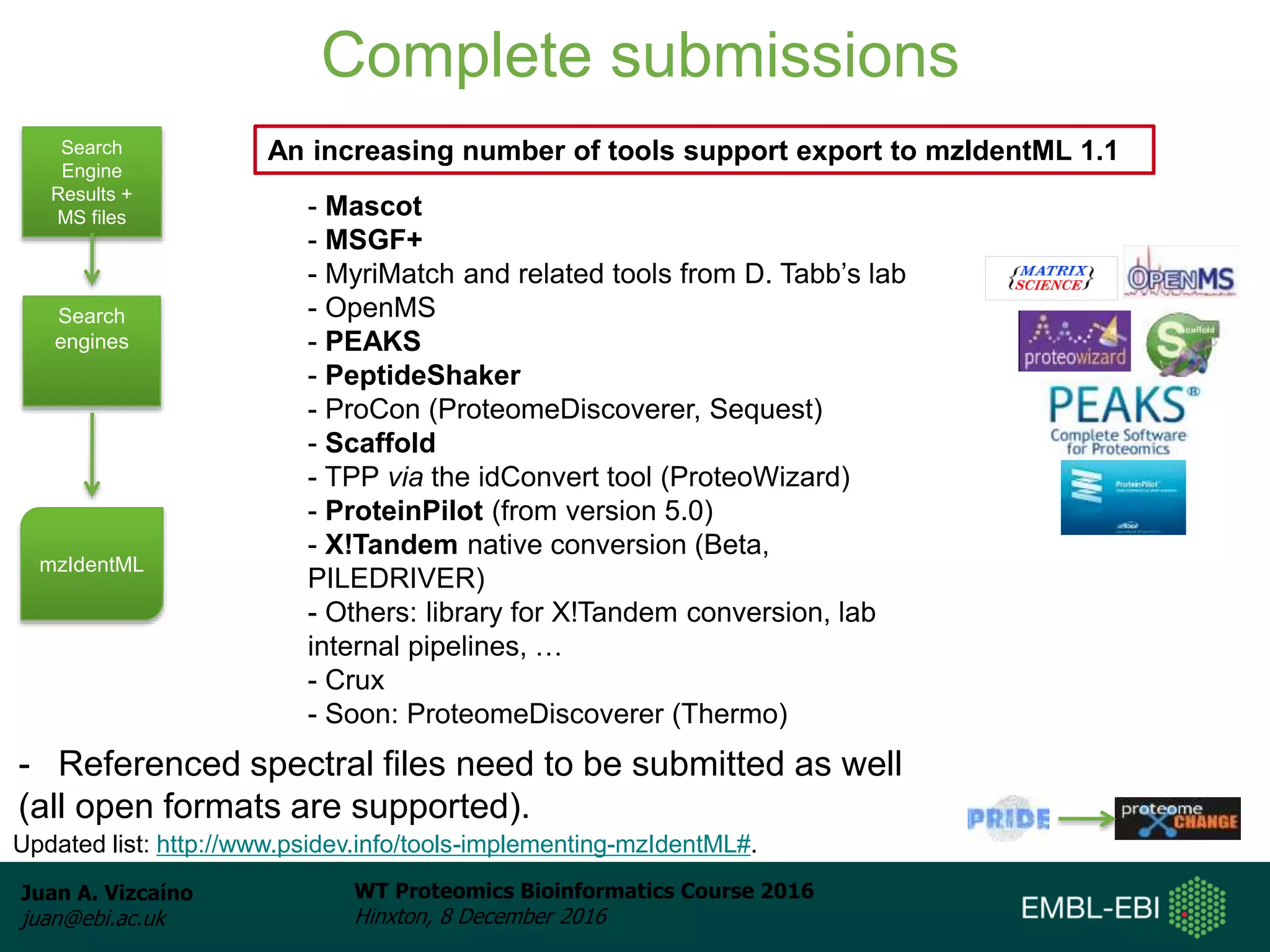
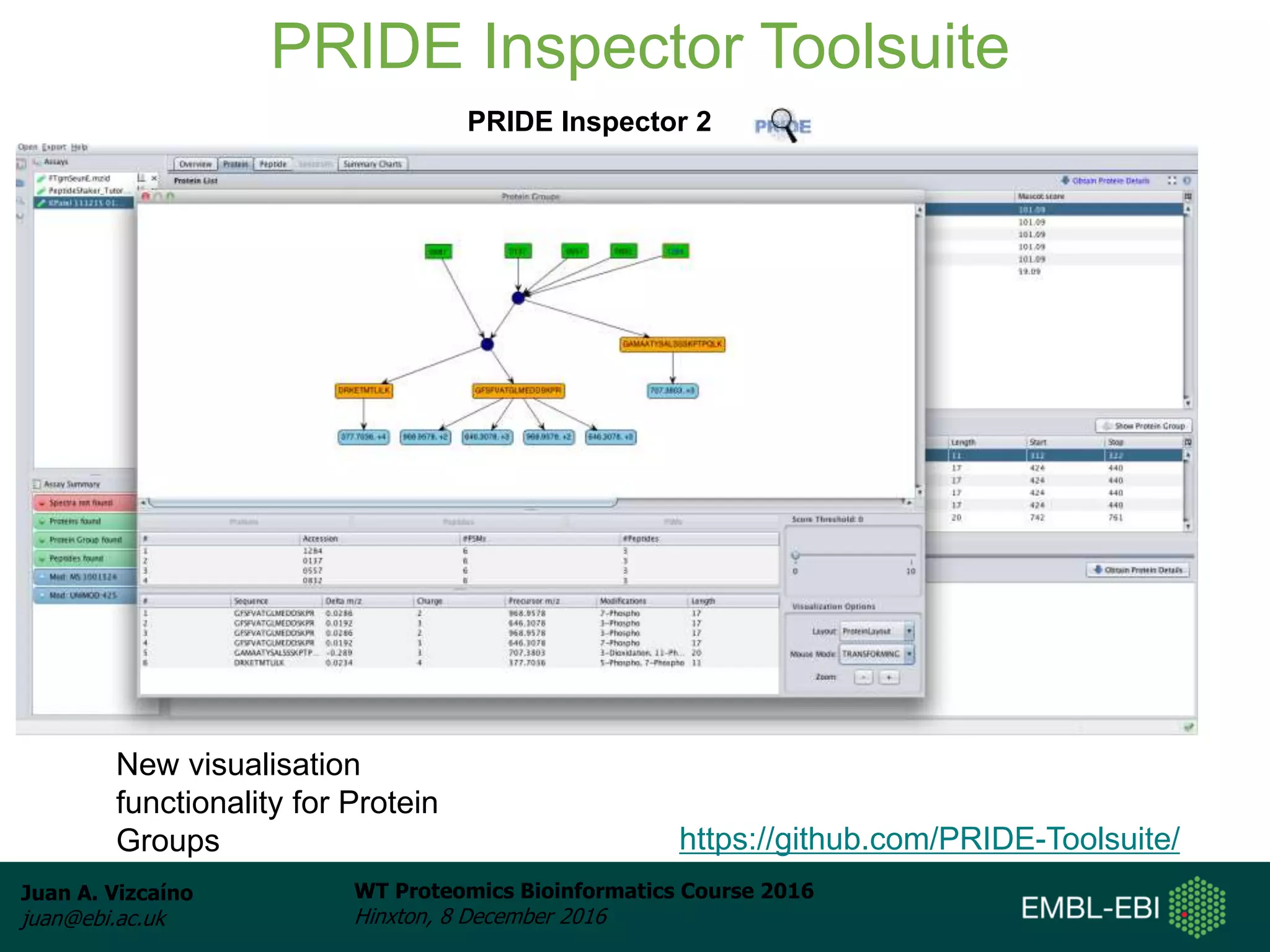
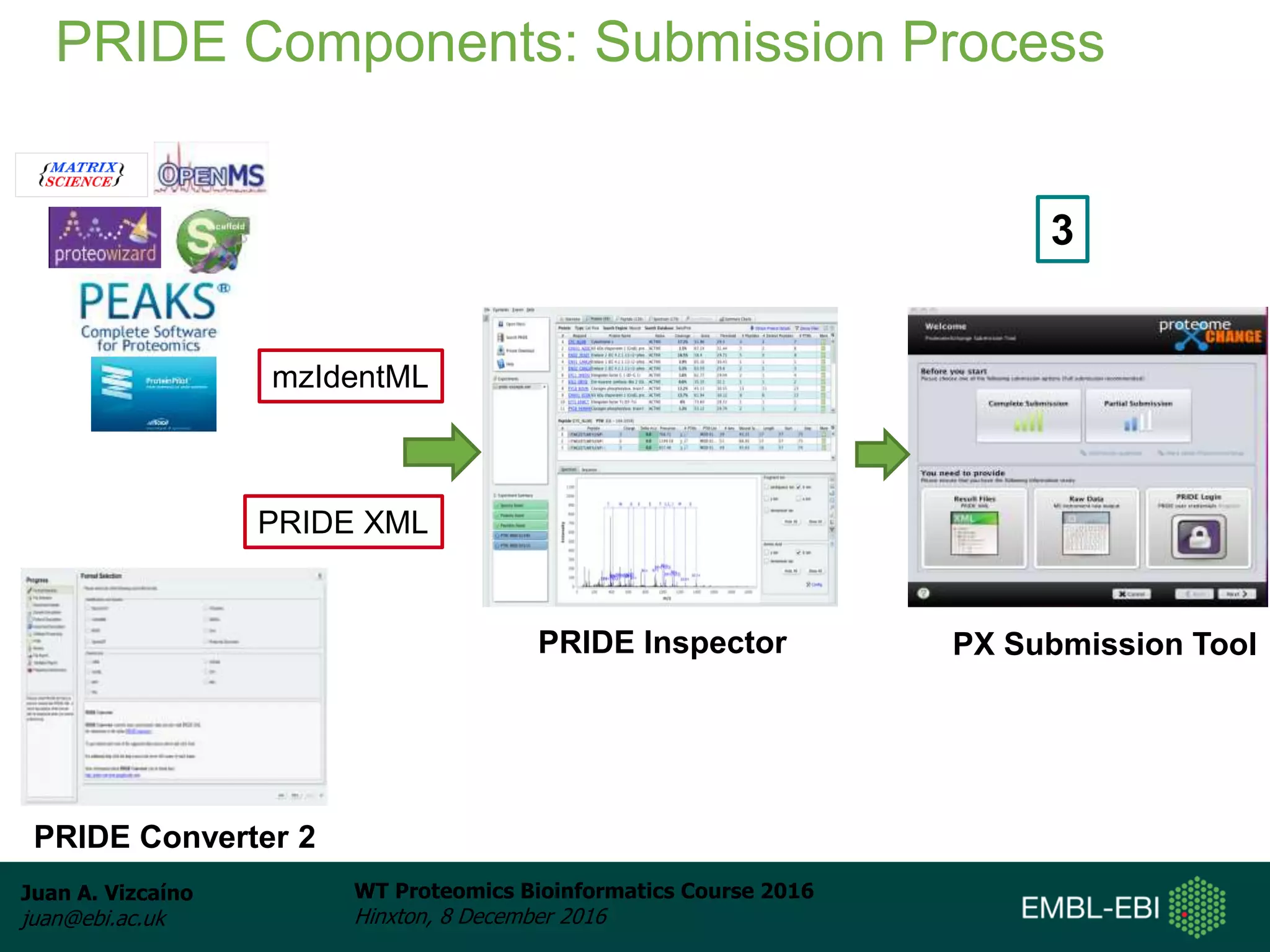
The document discusses PRIDE, a proteomics data repository at EMBL-EBI. It describes how PRIDE stores mass spectrometry proteomics data, its role within the ProteomeXchange consortium, and how researchers can submit data to PRIDE including the use of mzIdentML and PRIDE tools.



















![Juan A. Vizcaíno
juan@ebi.ac.uk
WT Proteomics Bioinformatics Course 2016
Hinxton, 8 December 2016
C
D
From original publication [13] Reconstructed ProteomeXchange data
1. Thermo RAW data / UDP
2. Mirion Software (JLU)
1. Thermo RAW data / UDP
2. Convert to imzML
3. Upload to PRIDE
(EBI, Cambridge, UK)
4. Download from PRIDE
5. Display in MSiReader
- Vendor-independent data format
- Freely available software (open source)
- ‘open data‘ – free to reuse
- Anybody can do this!
A public repository for mass spectrometry imaging data
Römpp et al., 2015
PRIDE
database
European
Bioinformatics
Institute,
Cambridge, UK
3. Upload
4. Download](https://image.slidesharecdn.com/03pridepx-161209110126/75/Pride-and-ProteomeXchange-31-2048.jpg)