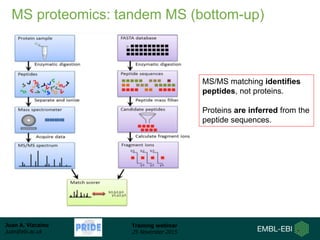
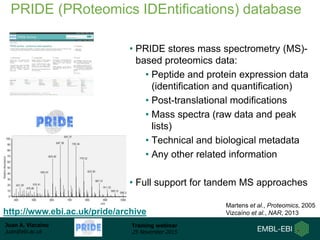
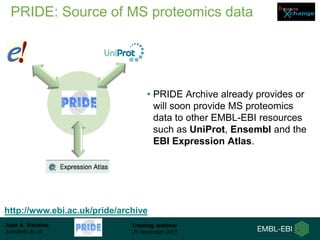
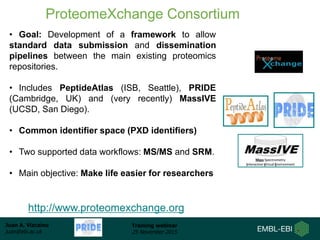
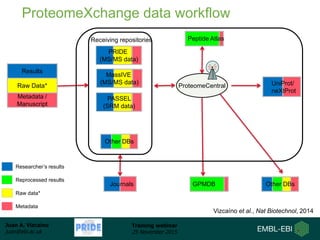
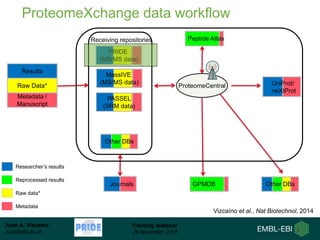
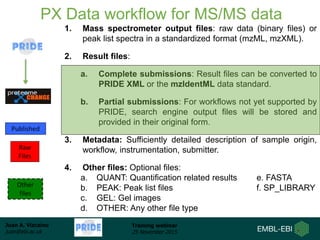
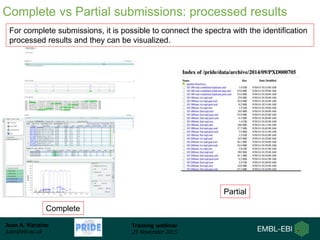
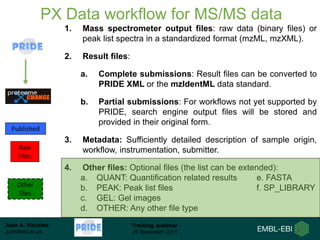
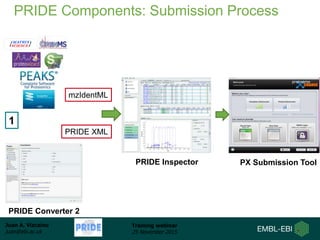
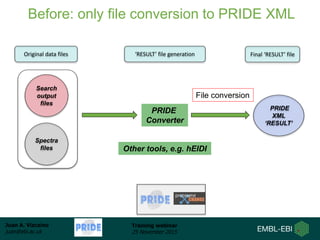
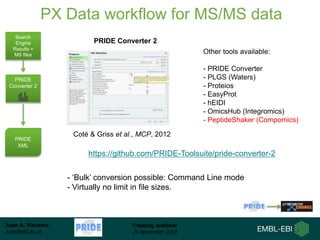
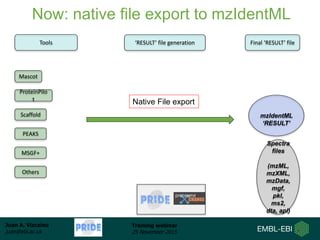
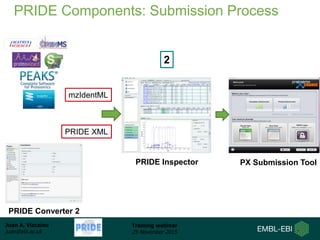
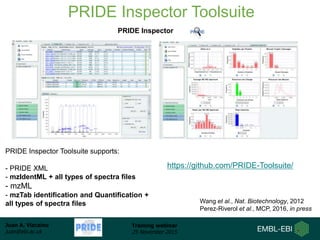
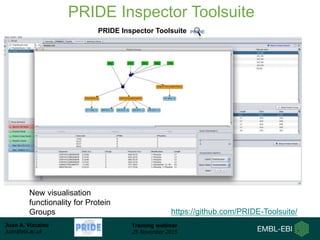
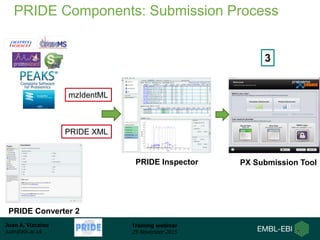
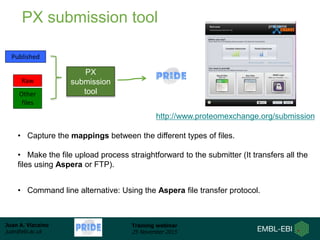
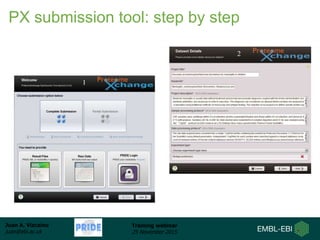
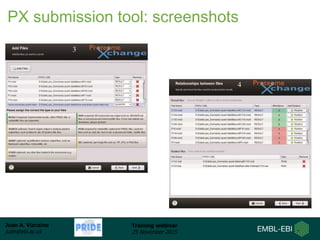
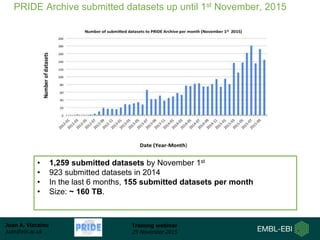
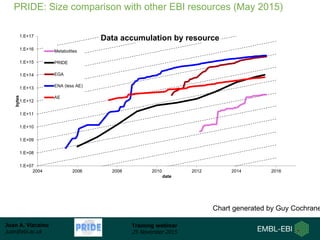
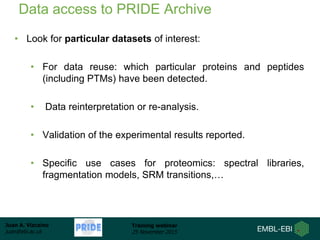
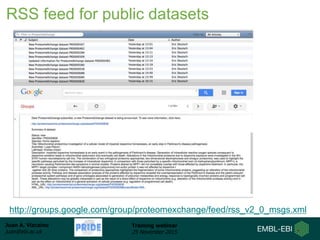
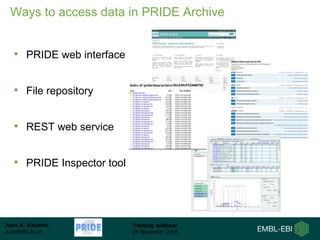
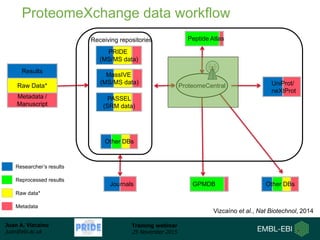
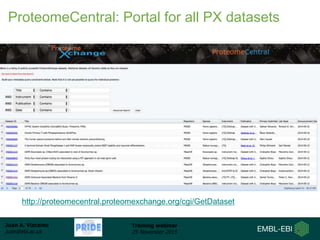
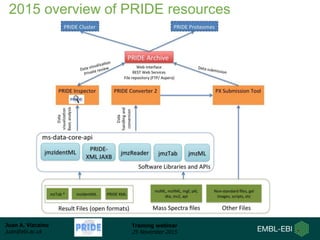
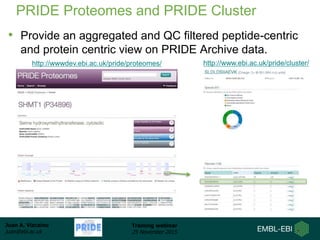
The document discusses a training webinar about PRIDE and ProteomeXchange. It begins with instructions for participating in the webinar and an overview of data resources at EMBL-EBI. It then covers PRIDE's mission to archive proteomics data, the ProteomeXchange consortium for standardized data submission, and tools for submitting data to PRIDE including PRIDE Converter, PRIDE Inspector, and the ProteomeXchange submission tool.