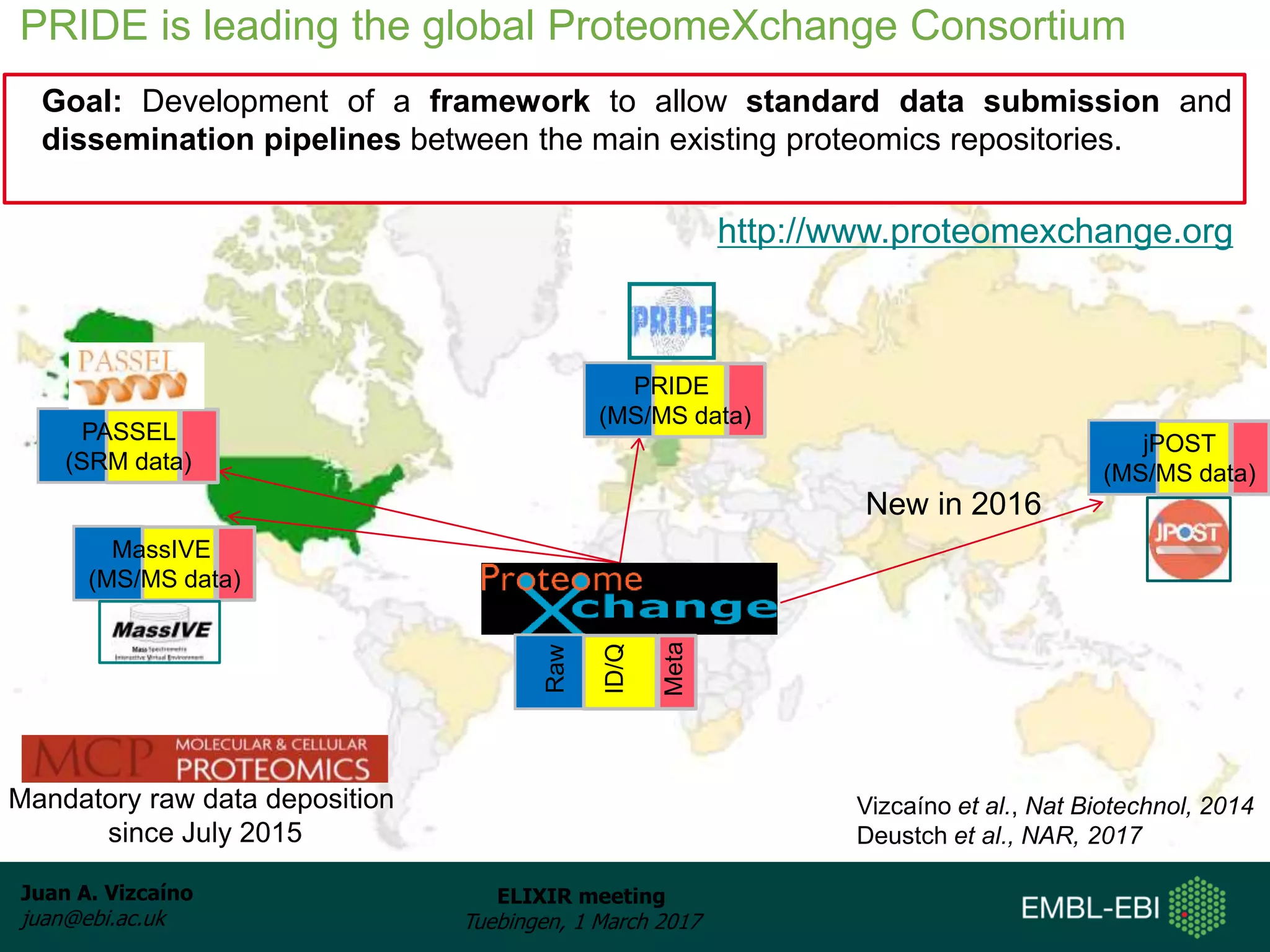
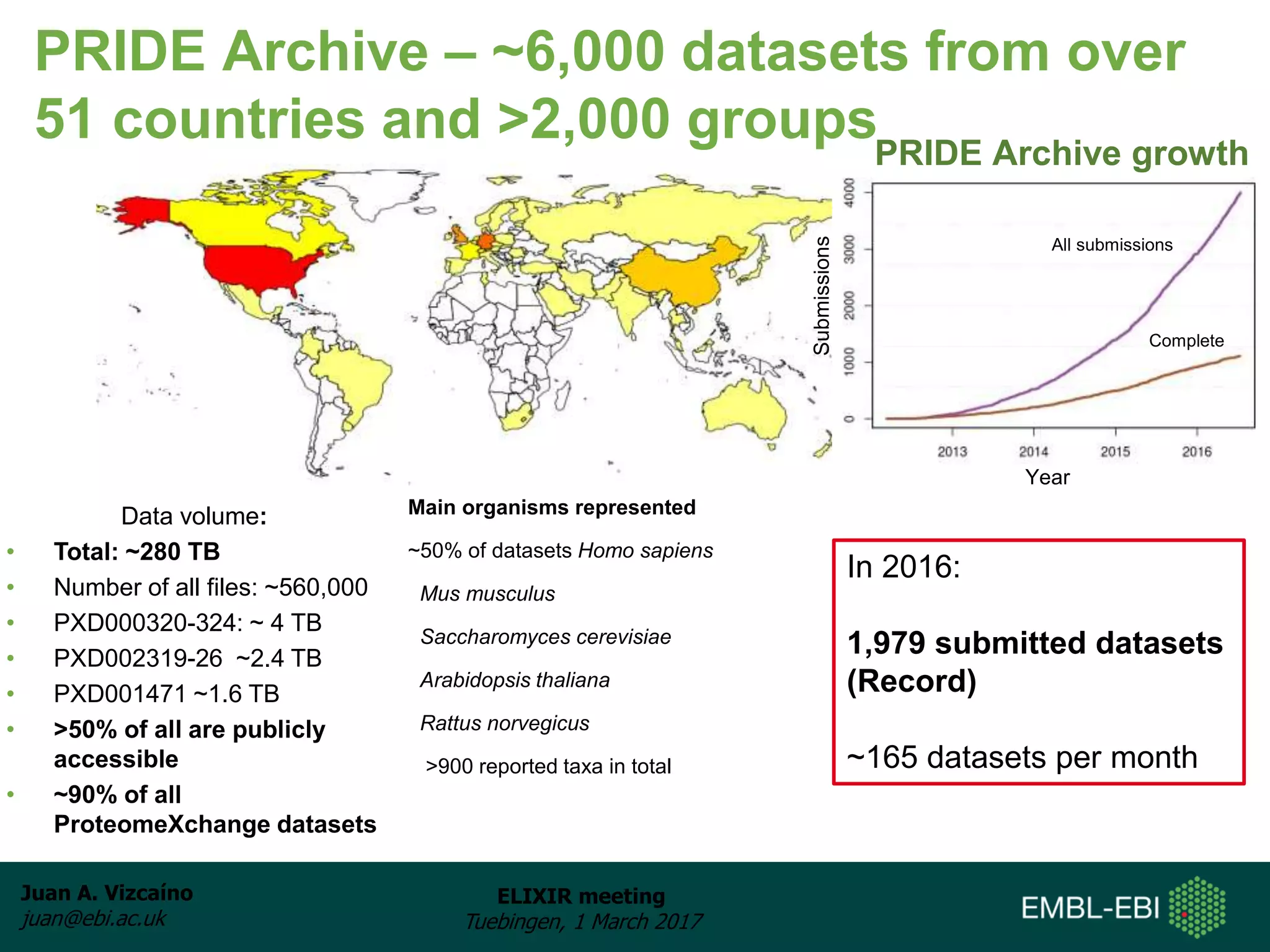
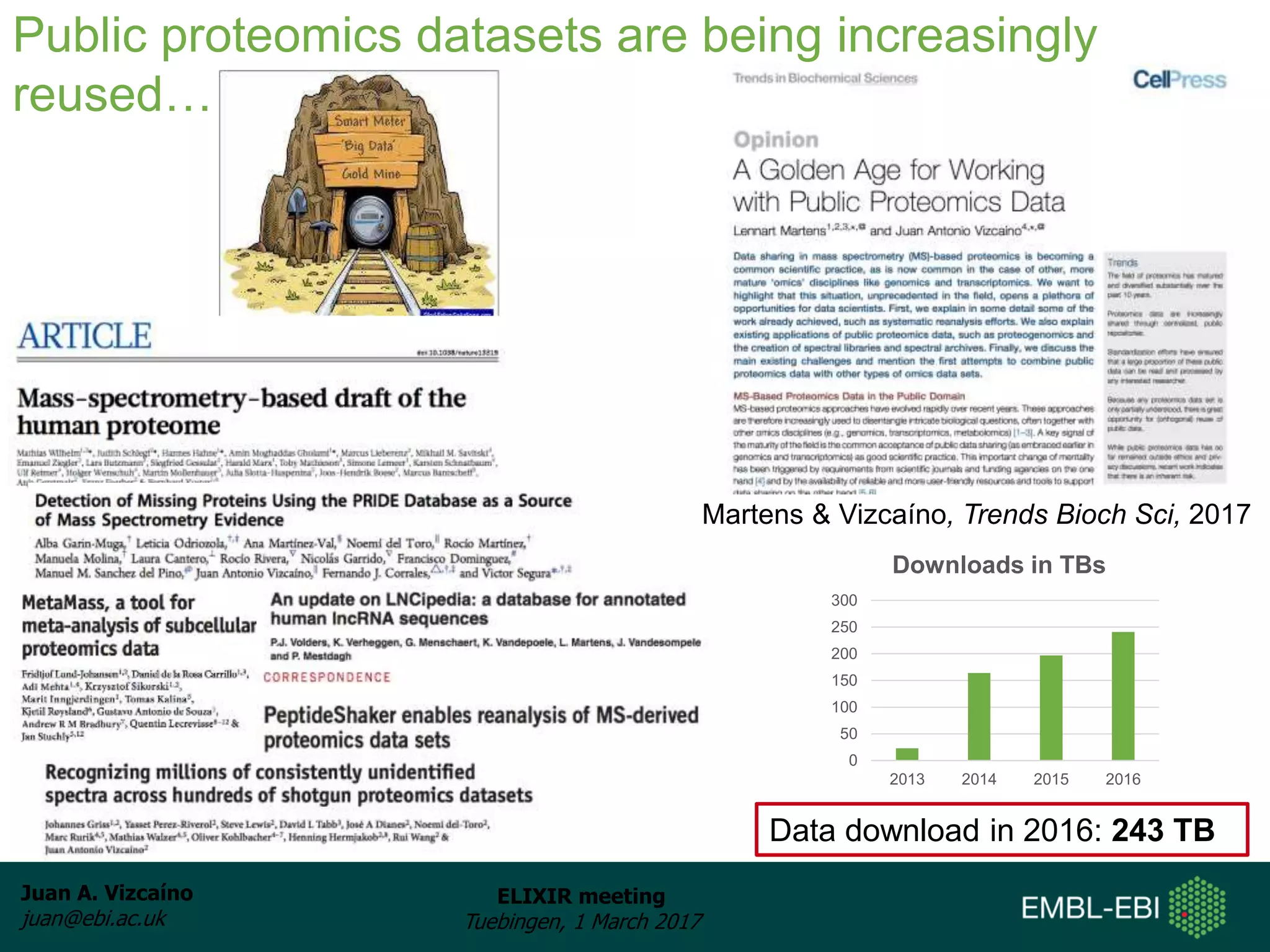
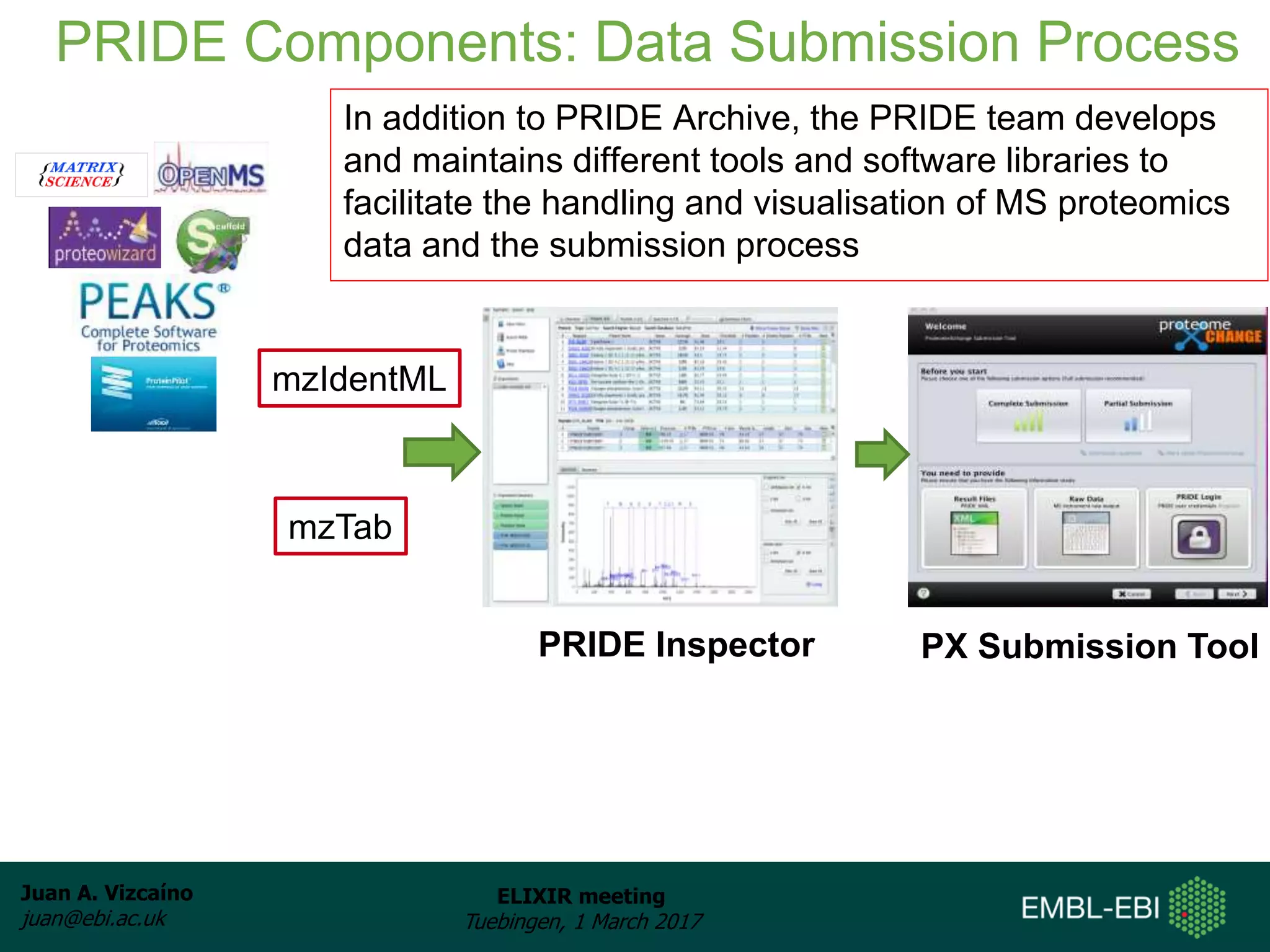
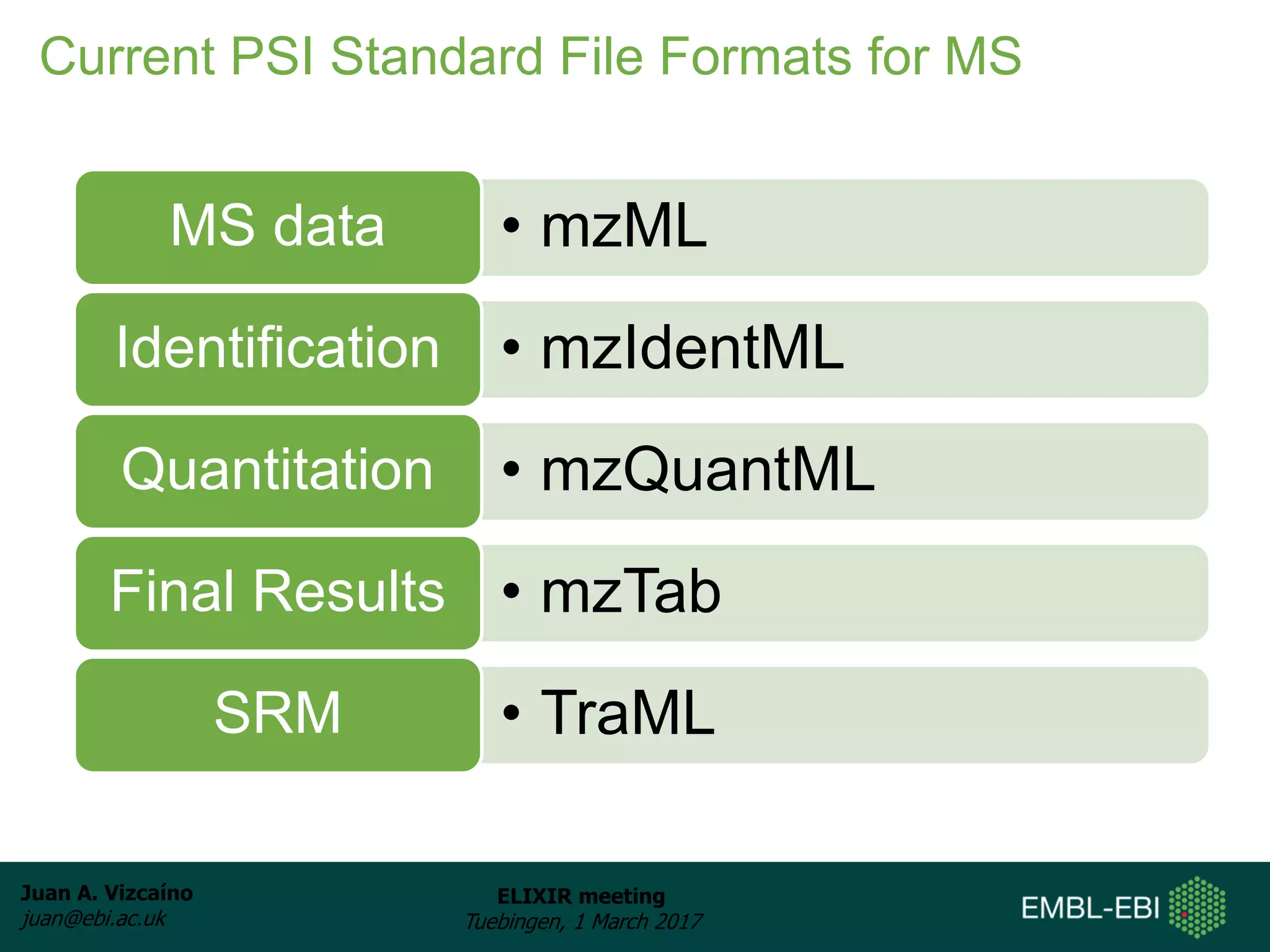
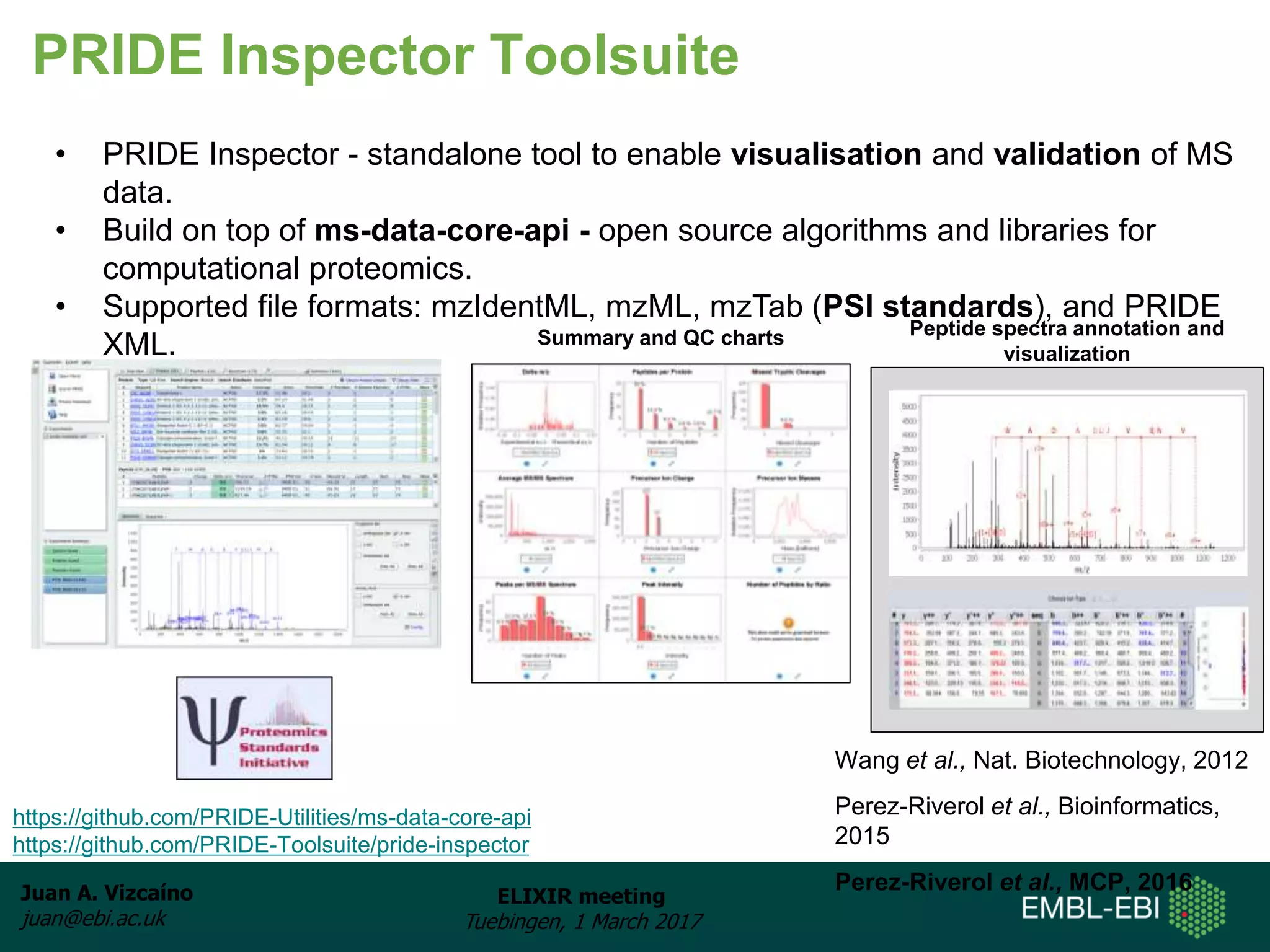
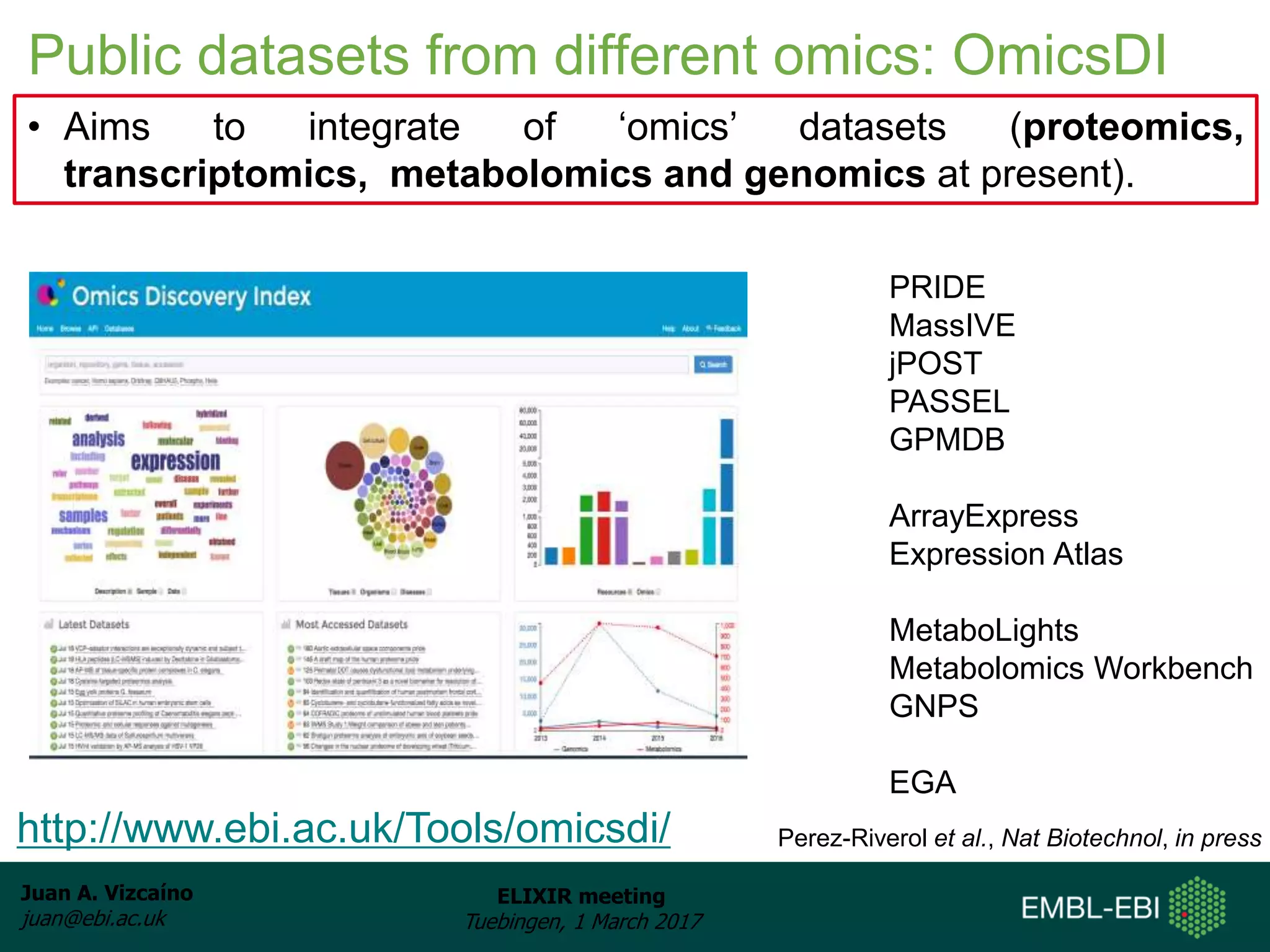
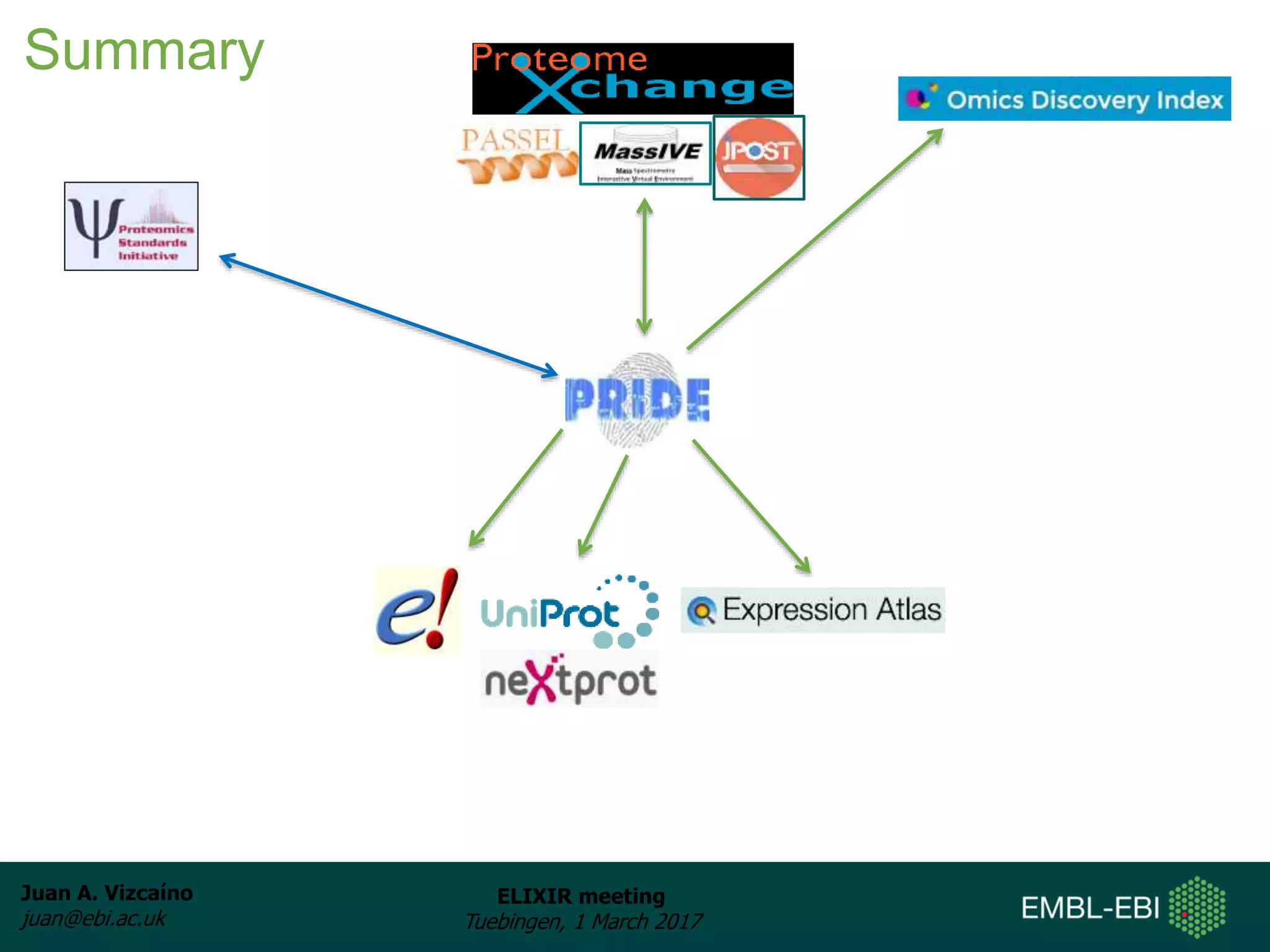
The document discusses the activities of the EMBL-EBI ELIXIR Node related to proteomics data and analysis. It describes how EMBL-EBI contributes to the ELIXIR platforms of data, tools, interoperability, compute, and training through its work on the PRIDE Archive and ProteomeXchange repository, development of proteomics data standards and software tools, implementation of reproducible proteomics pipelines, and proteomics training courses. The PRIDE Archive contains over 280 terabytes of mass spectrometry proteomics data from over 51 countries and has seen rapid growth in recent years.