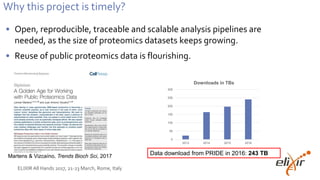
This document summarizes several presentations and events related to proteomics data analysis and ELIXIR activities. It describes a kickoff meeting in Tuebingen where 25 people from 11 ELIXIR nodes discussed future proteomics activities. It also outlines a new 1-year ELIXIR implementation project led by EMBL-EBI and ELIXIR-Germany to develop reusable proteomics analysis pipelines using the OpenMS framework and deploy them on the EMBL-EBI cloud for processing large proteomics datasets from the PRIDE repository, which saw over 243 terabytes of data downloaded in 2016.