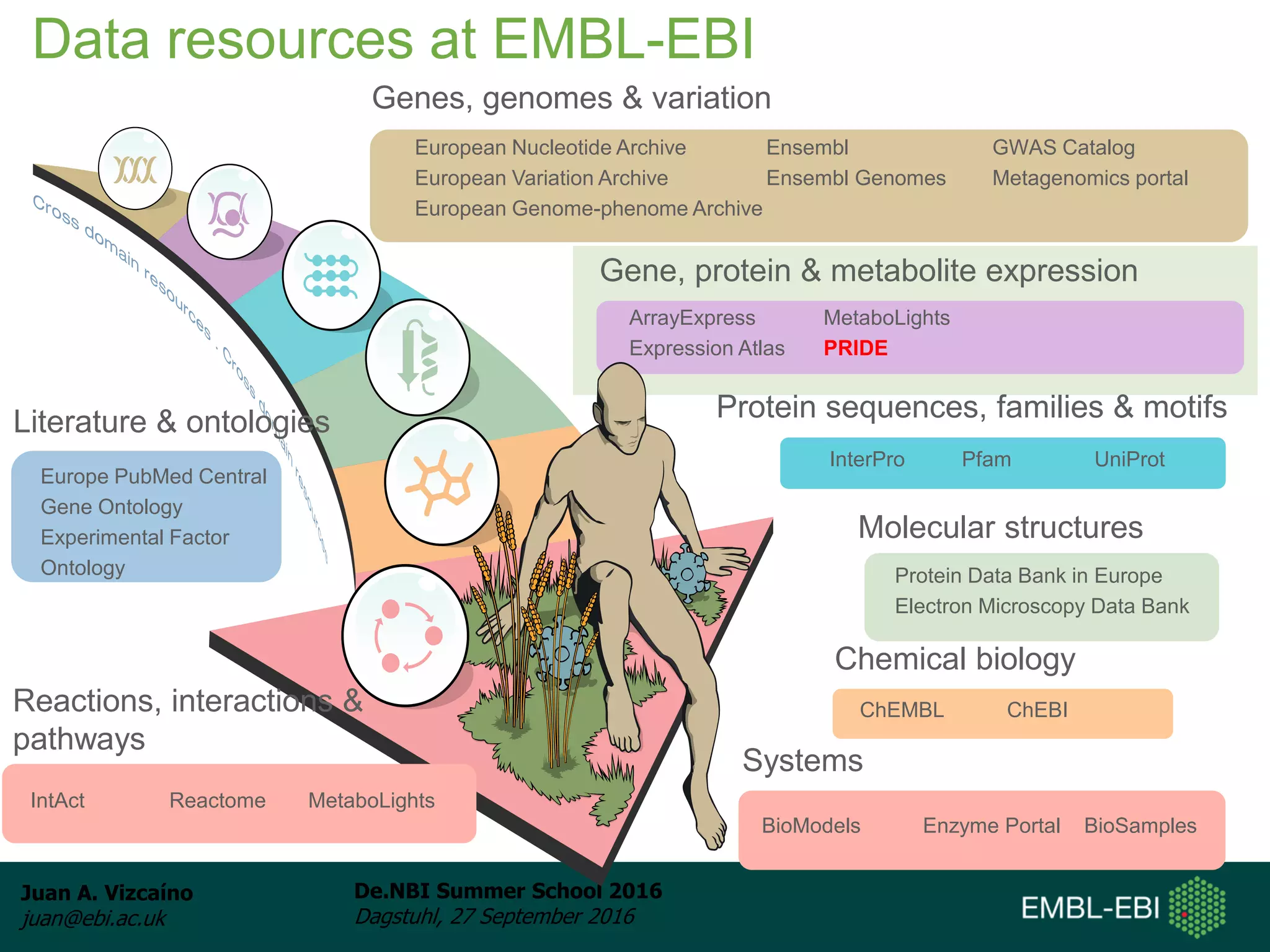
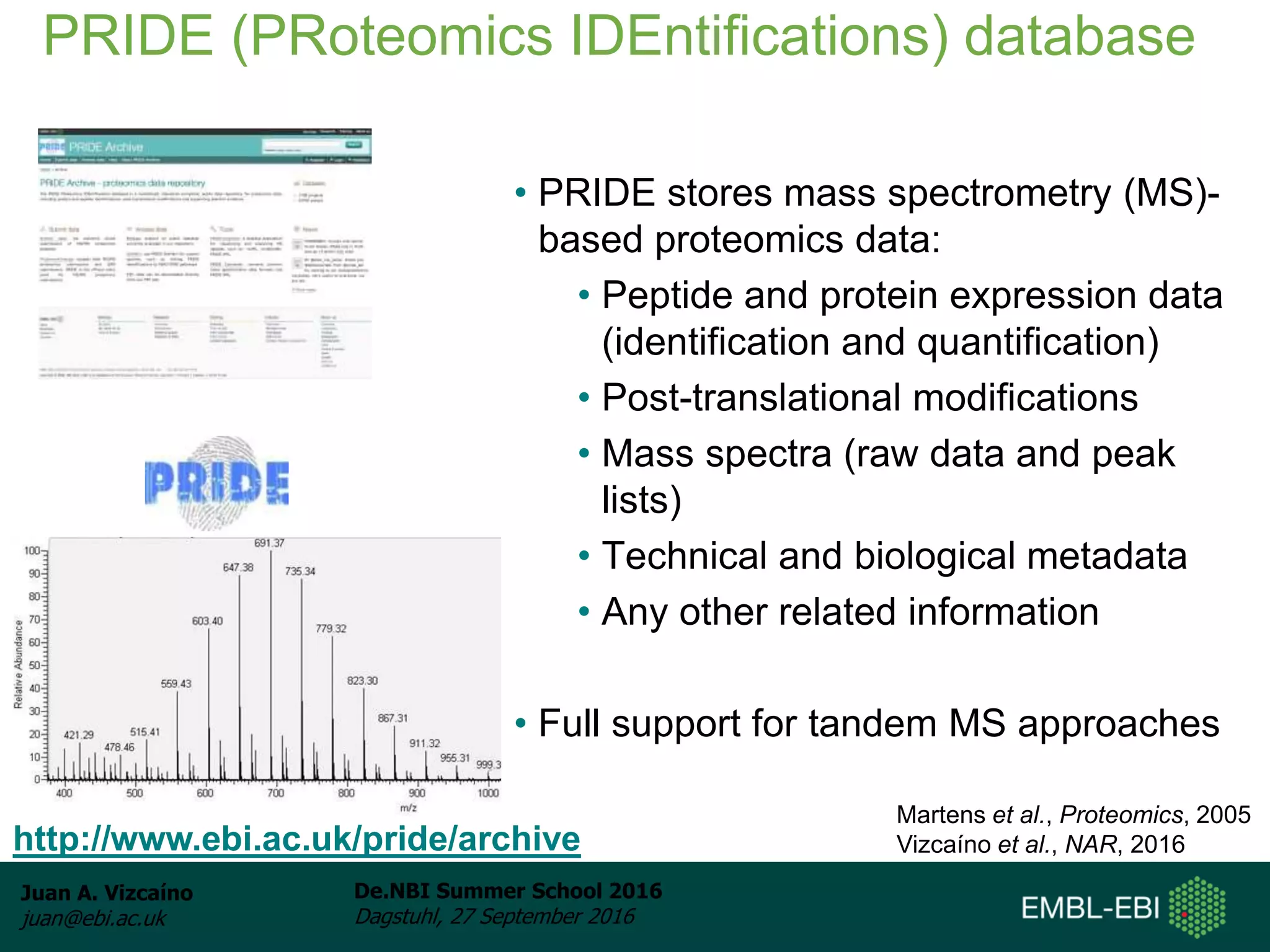
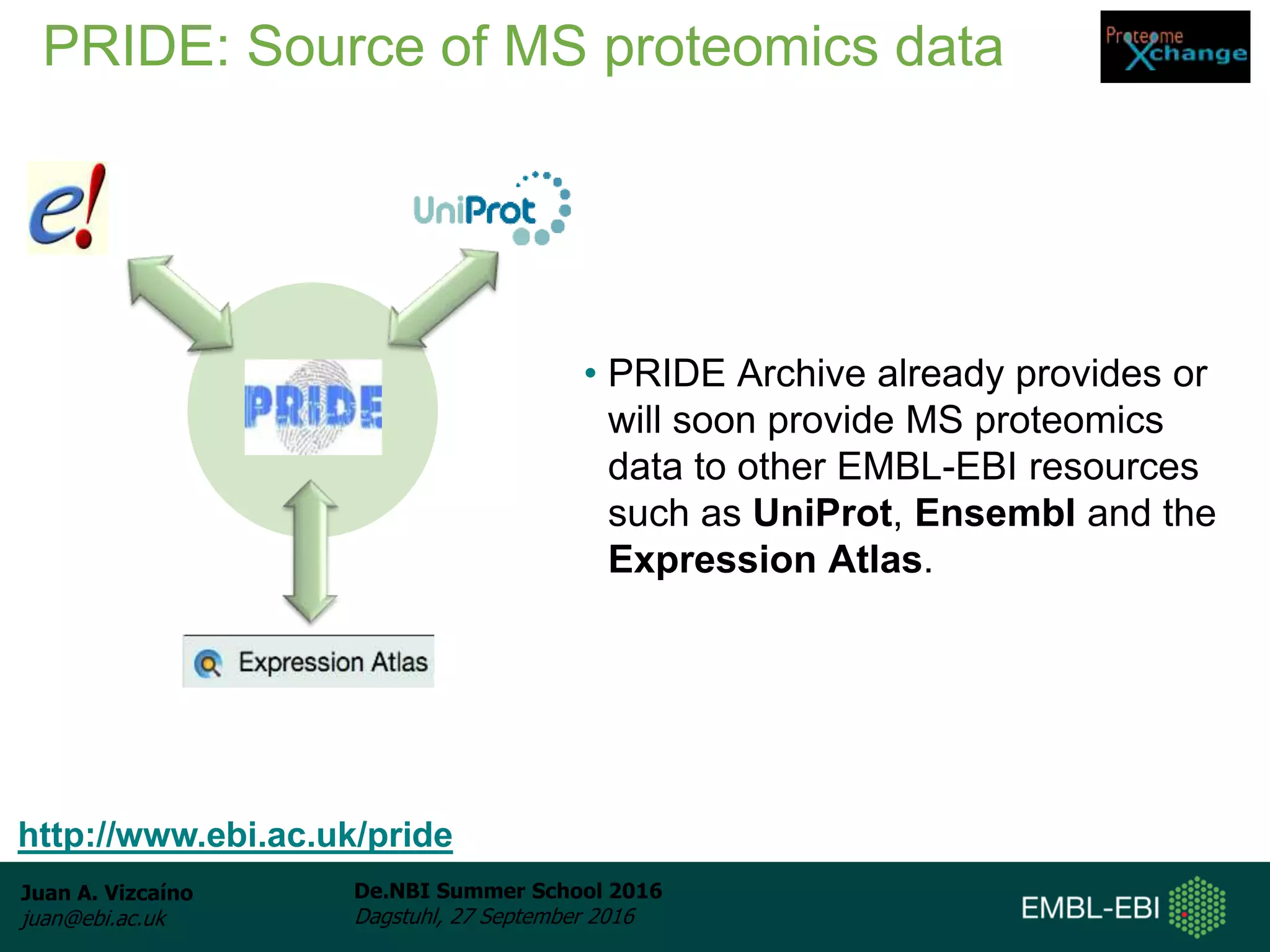
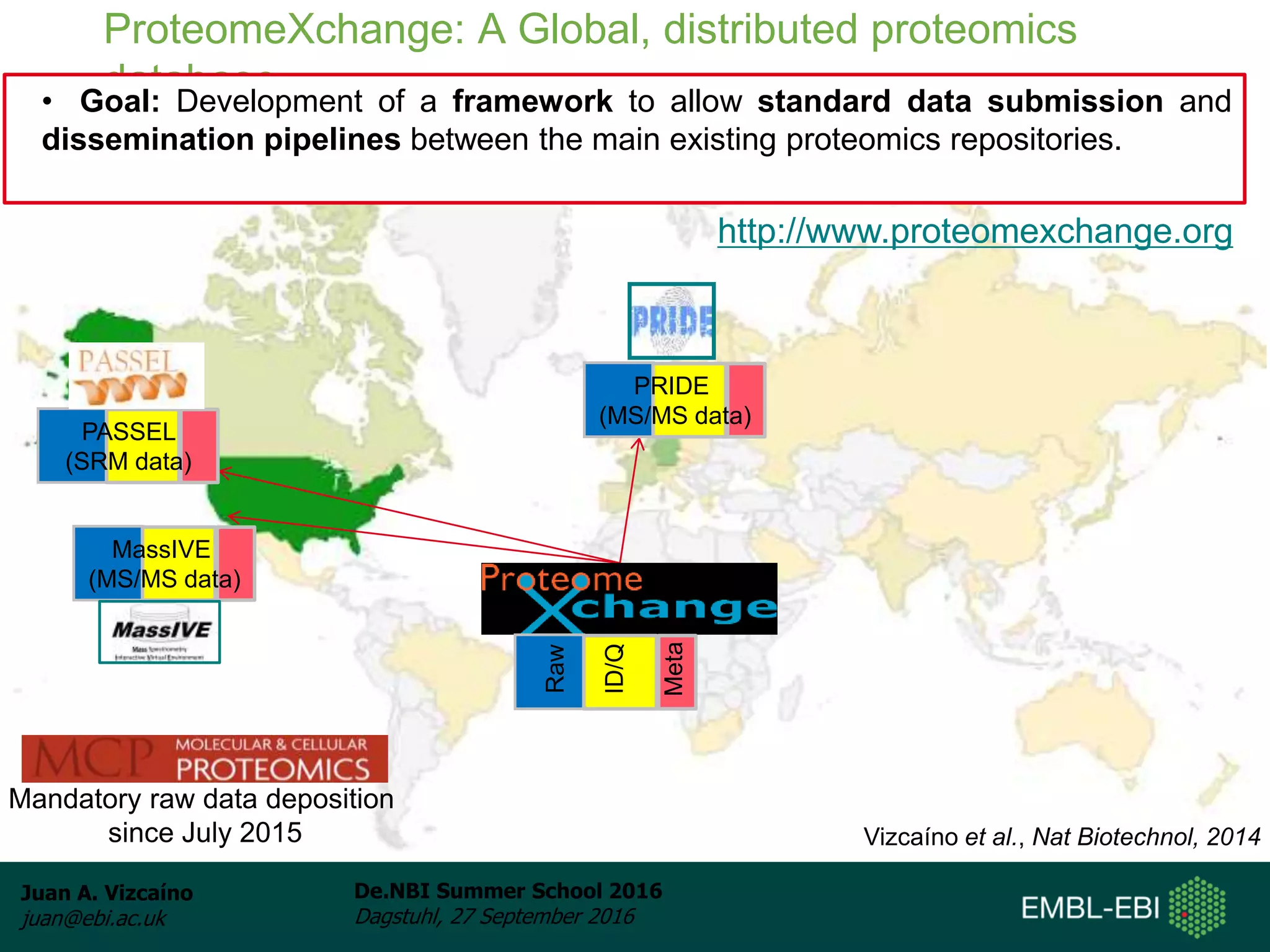
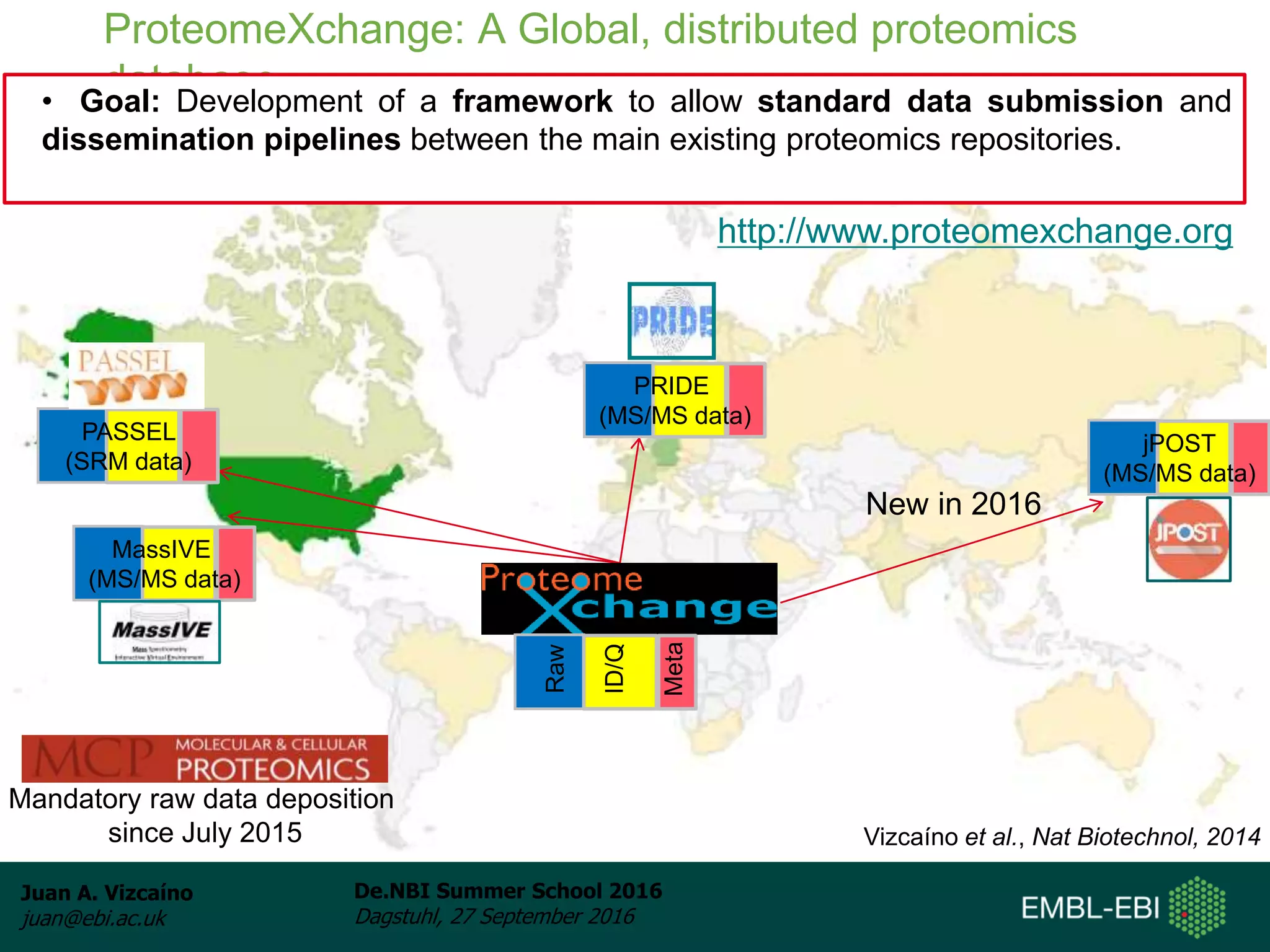
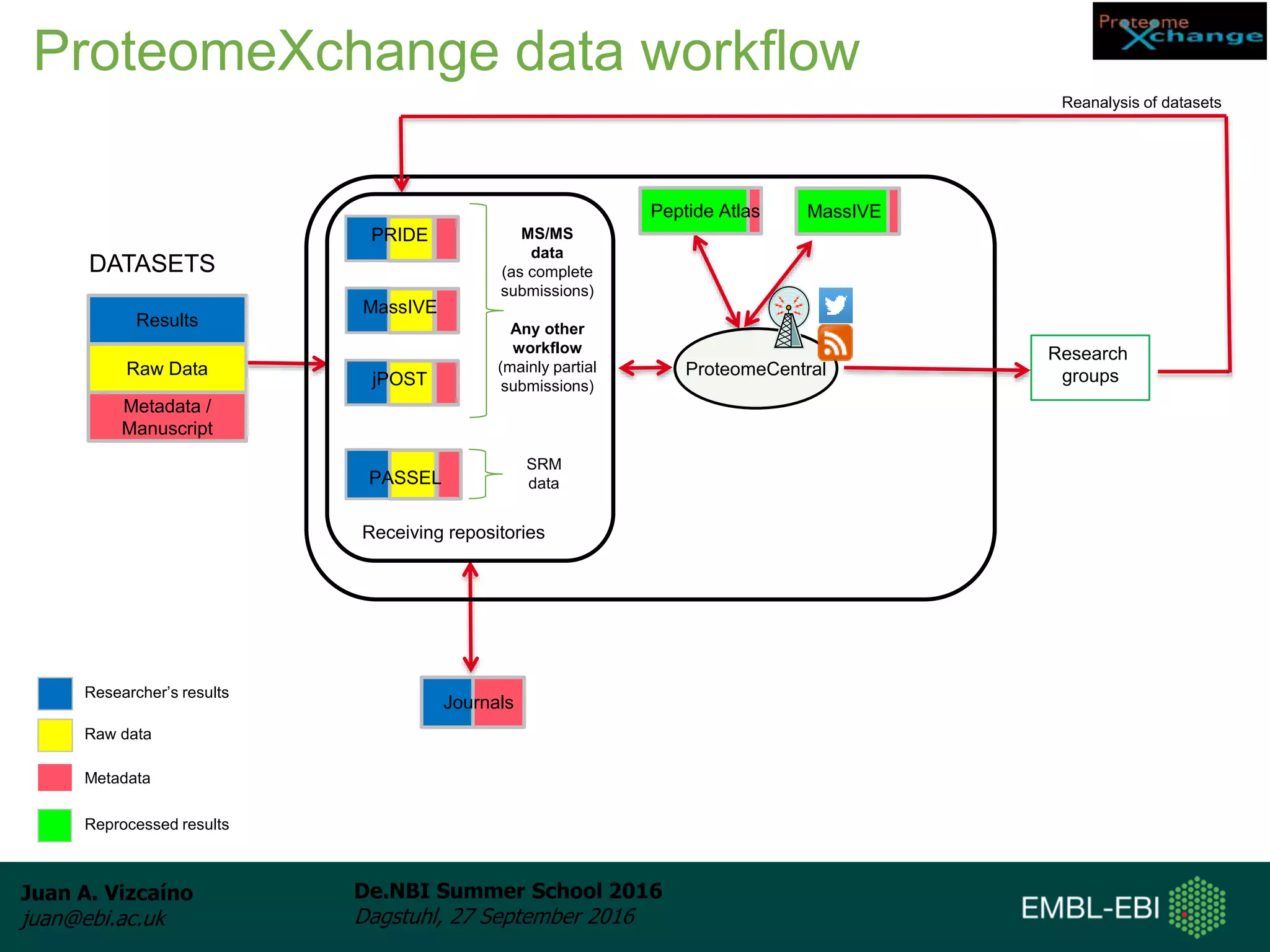
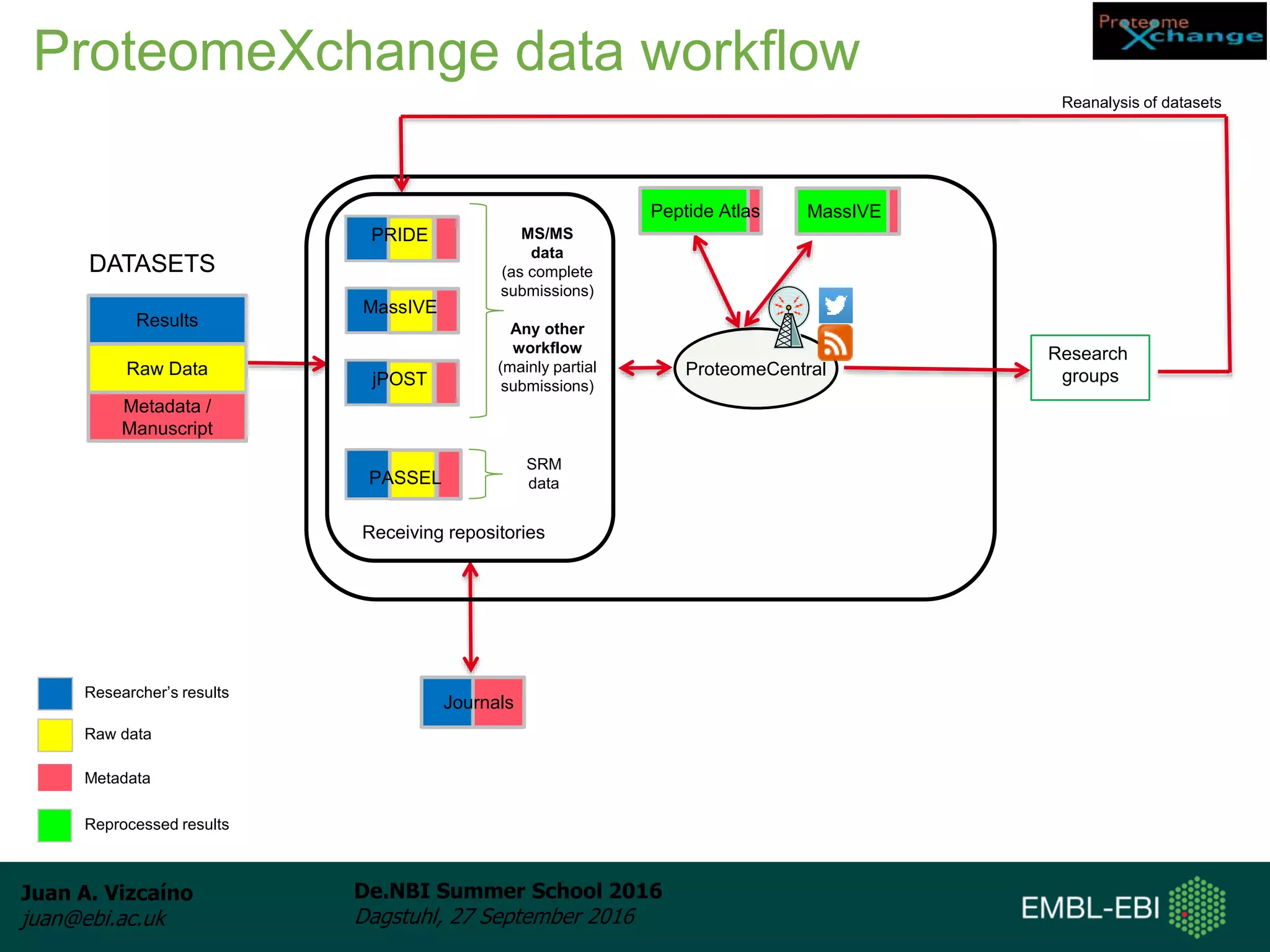
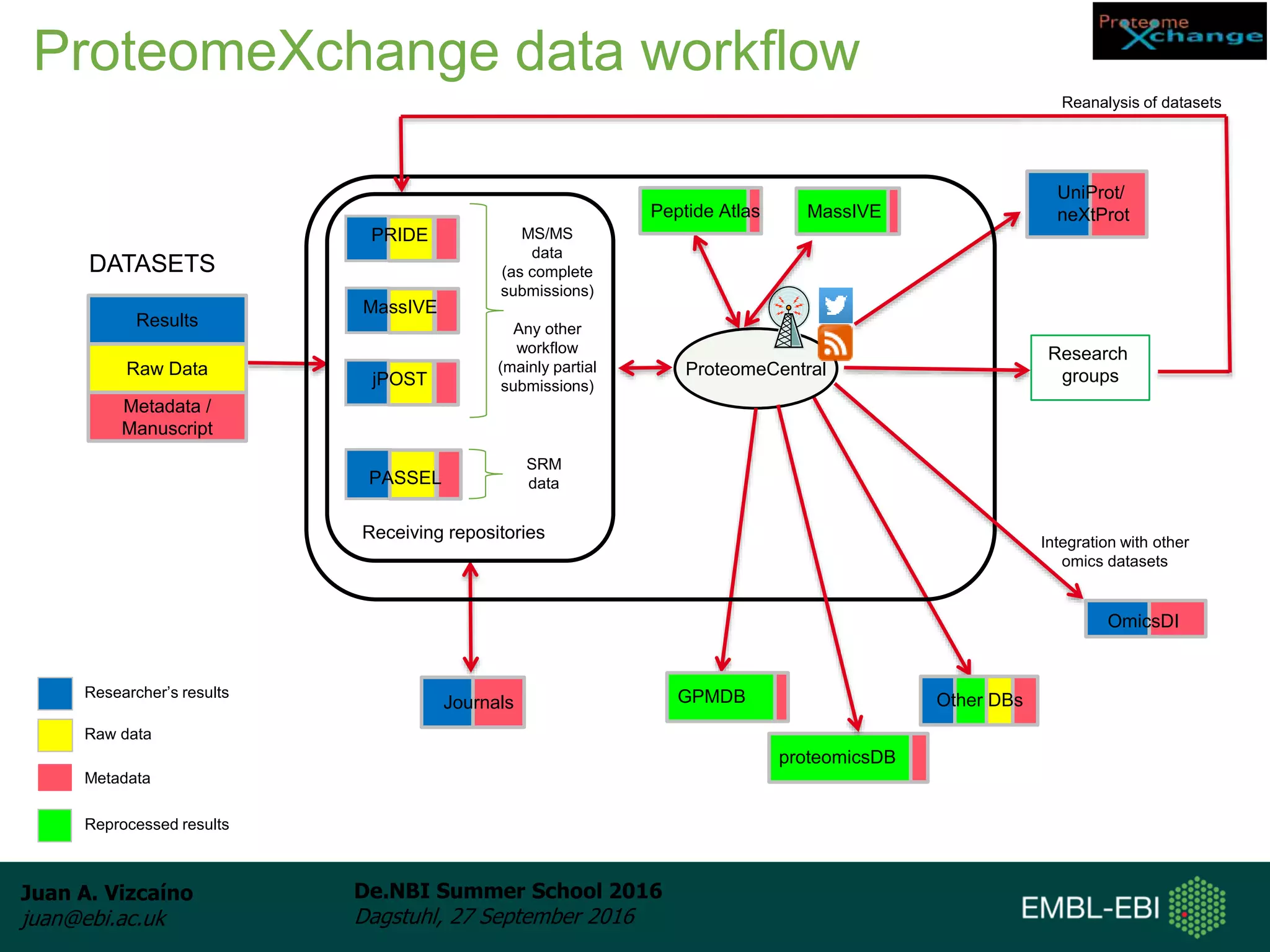
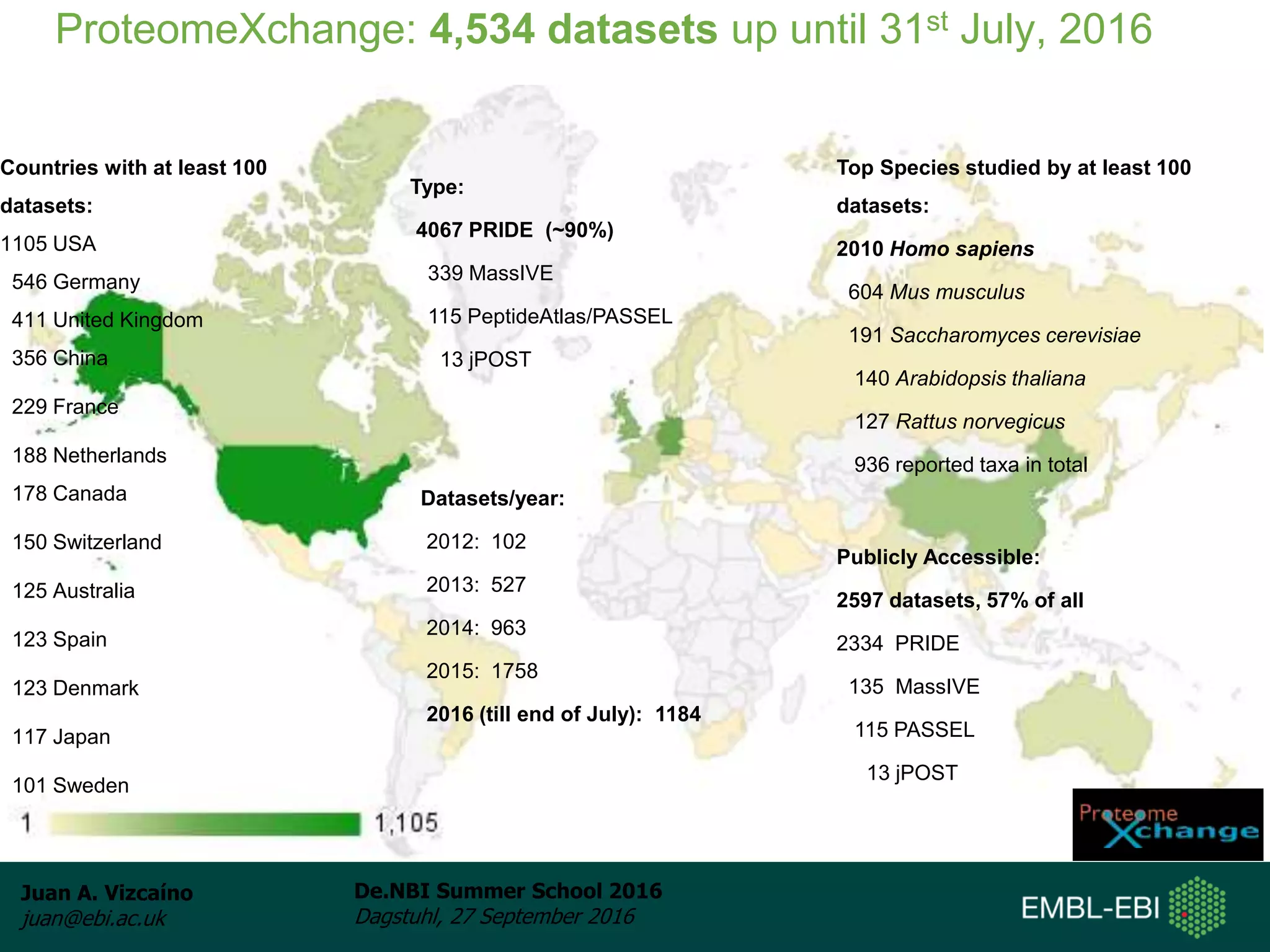
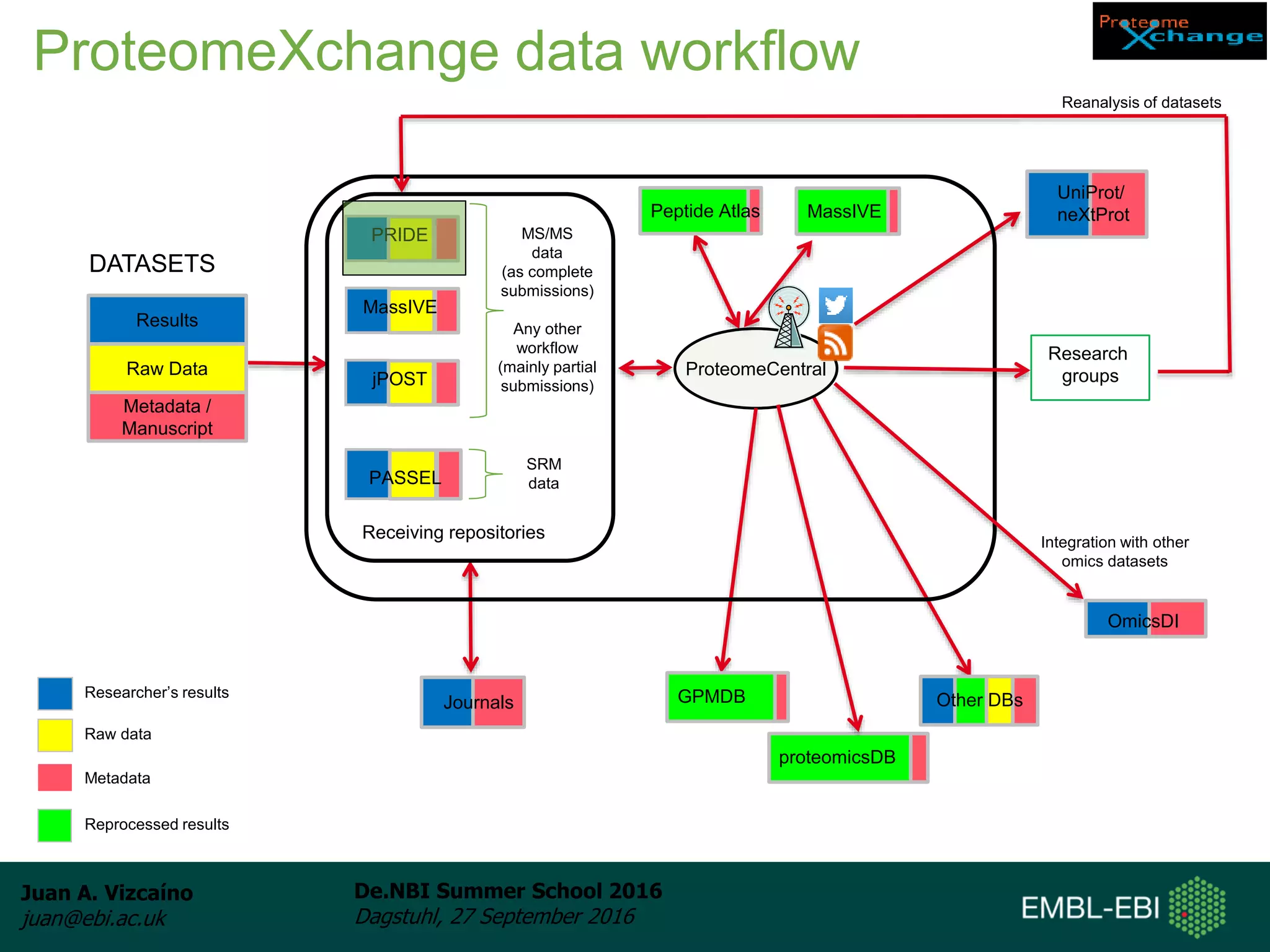
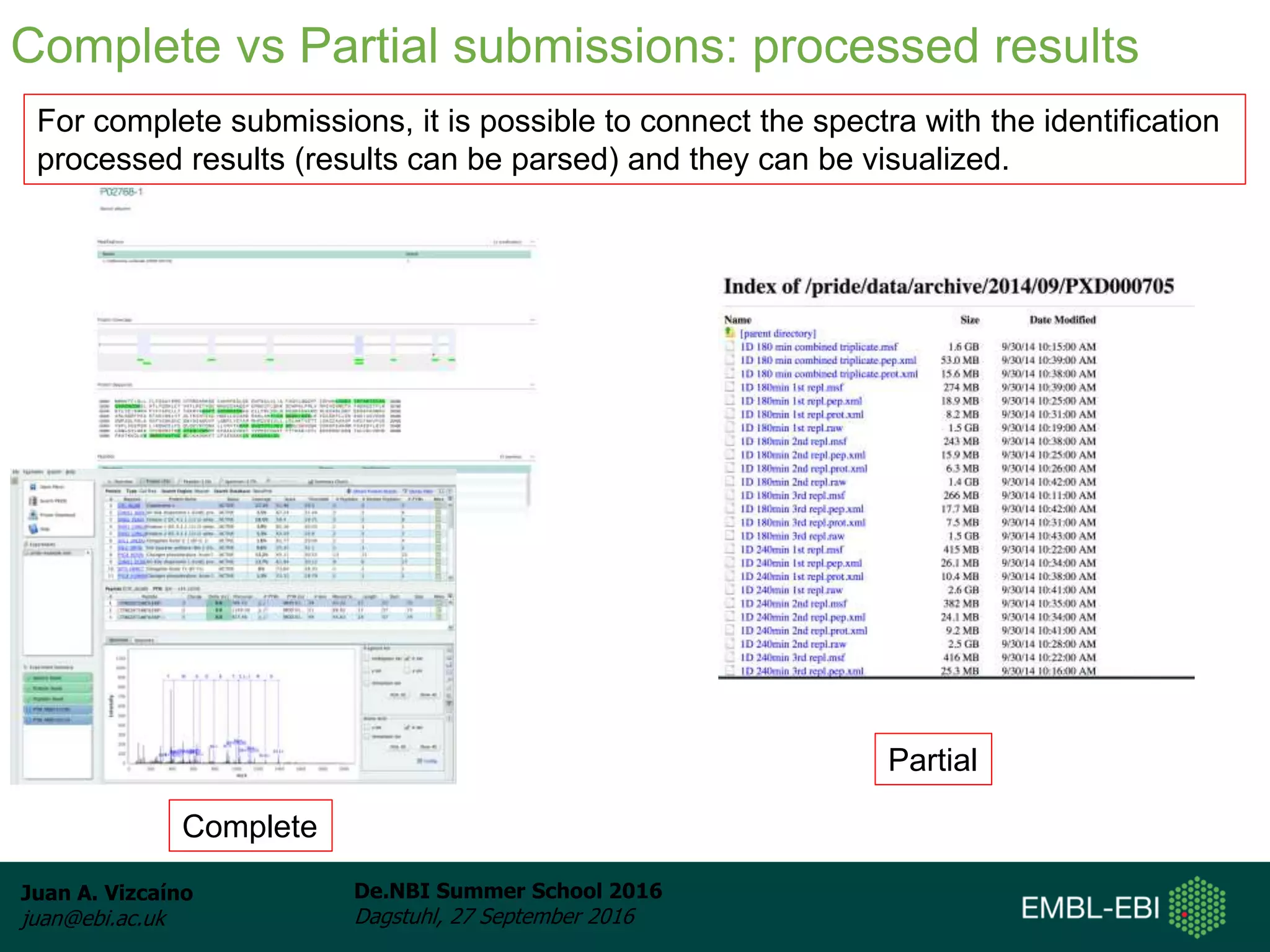
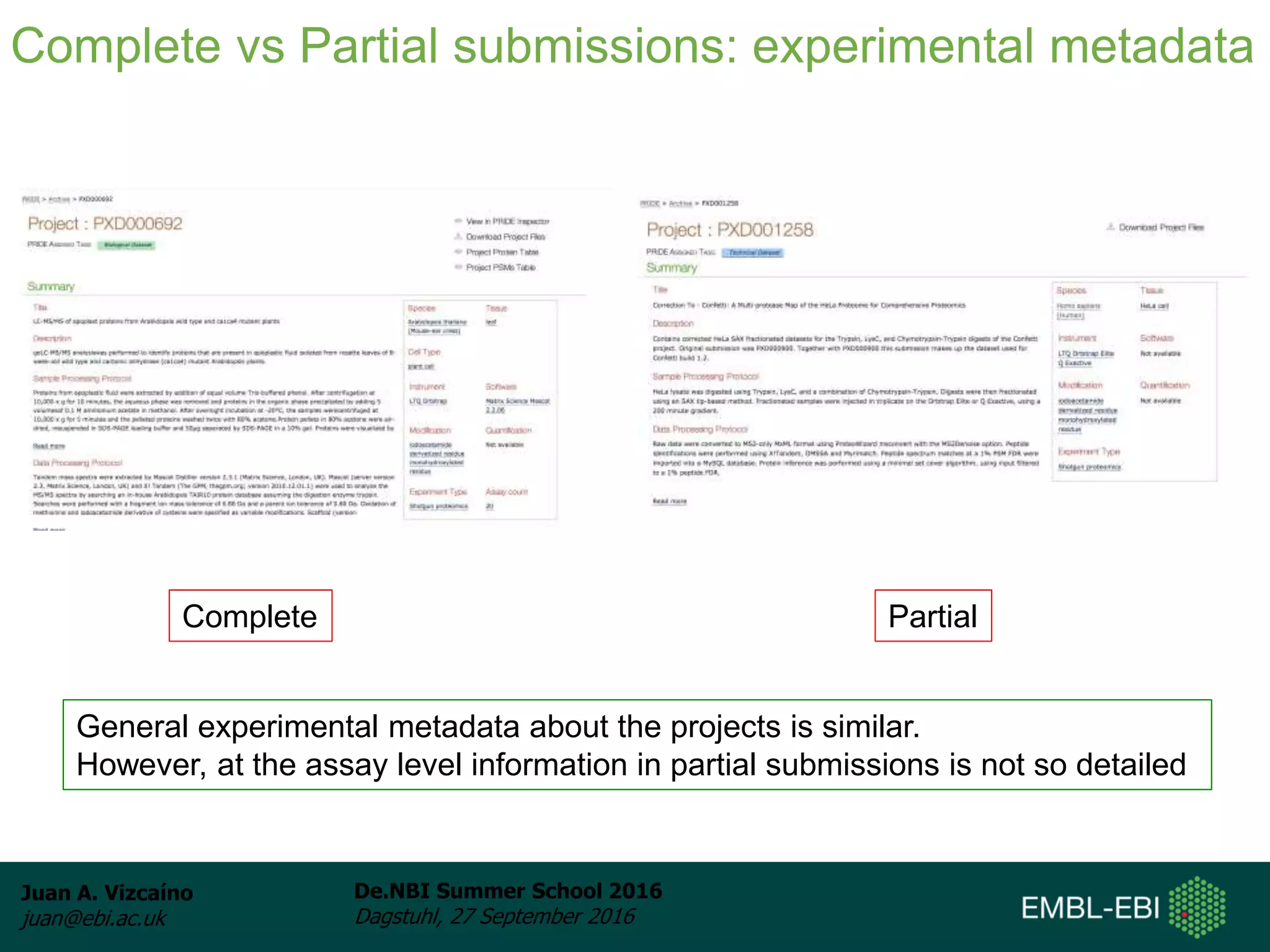
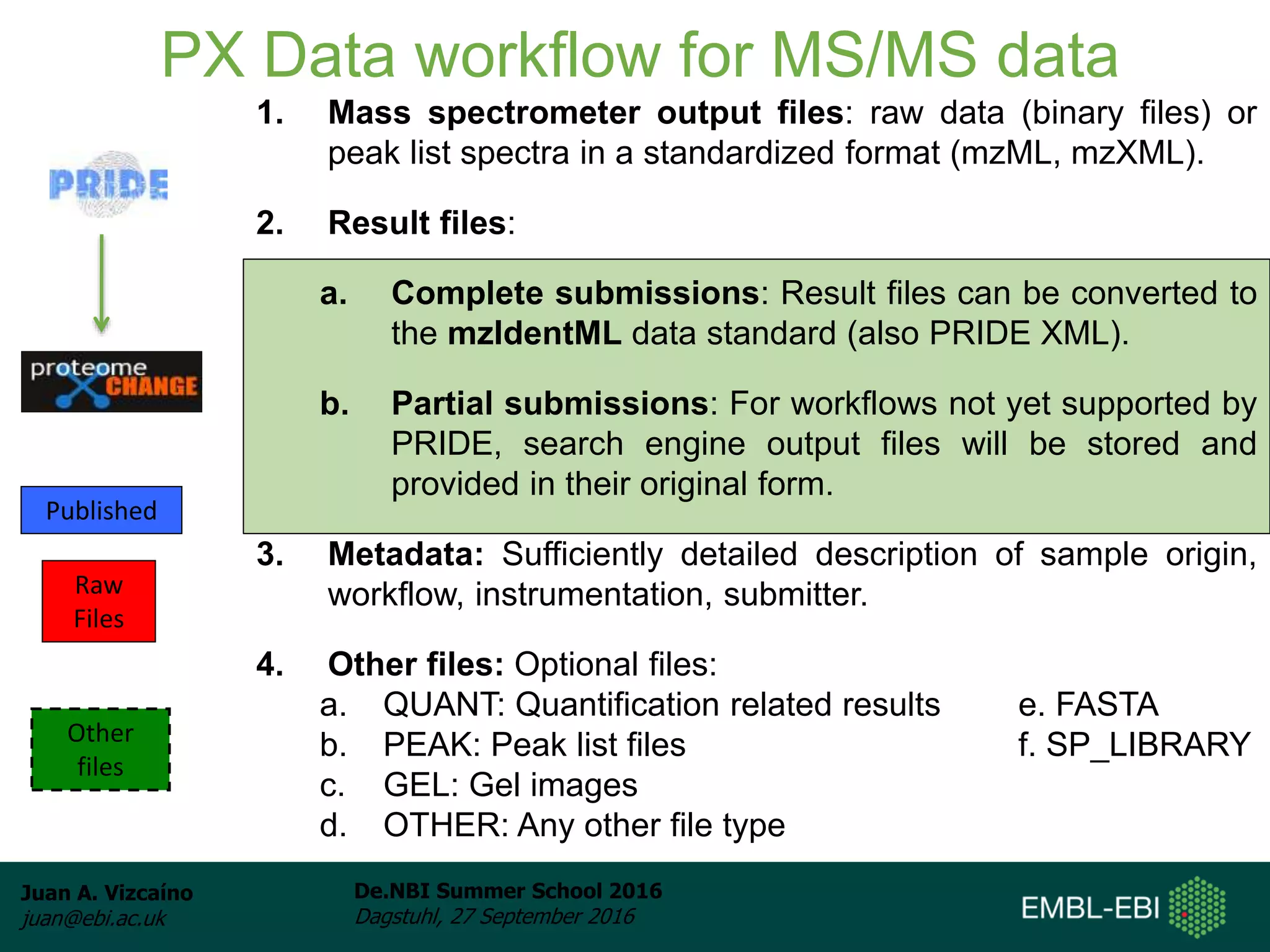
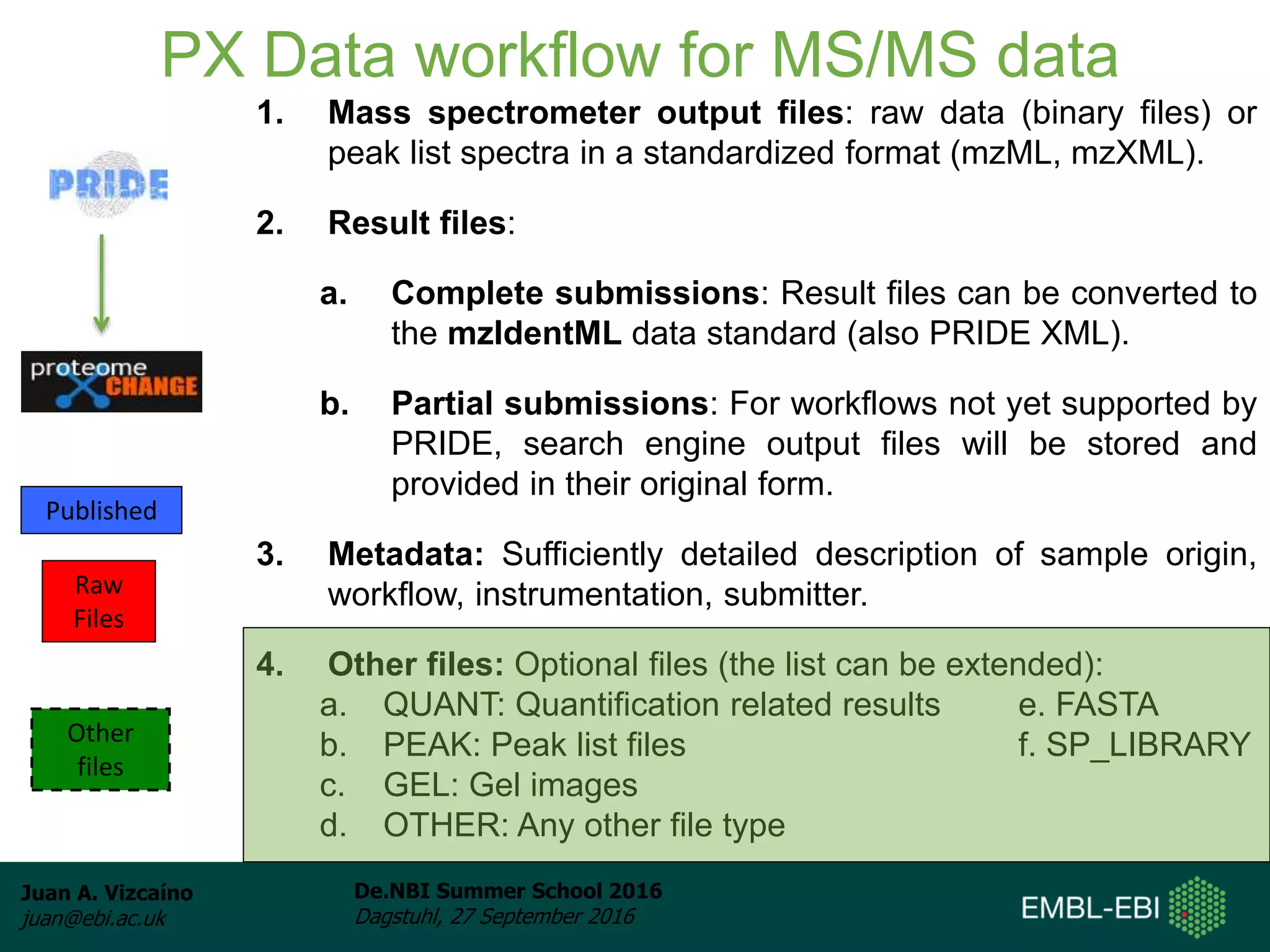
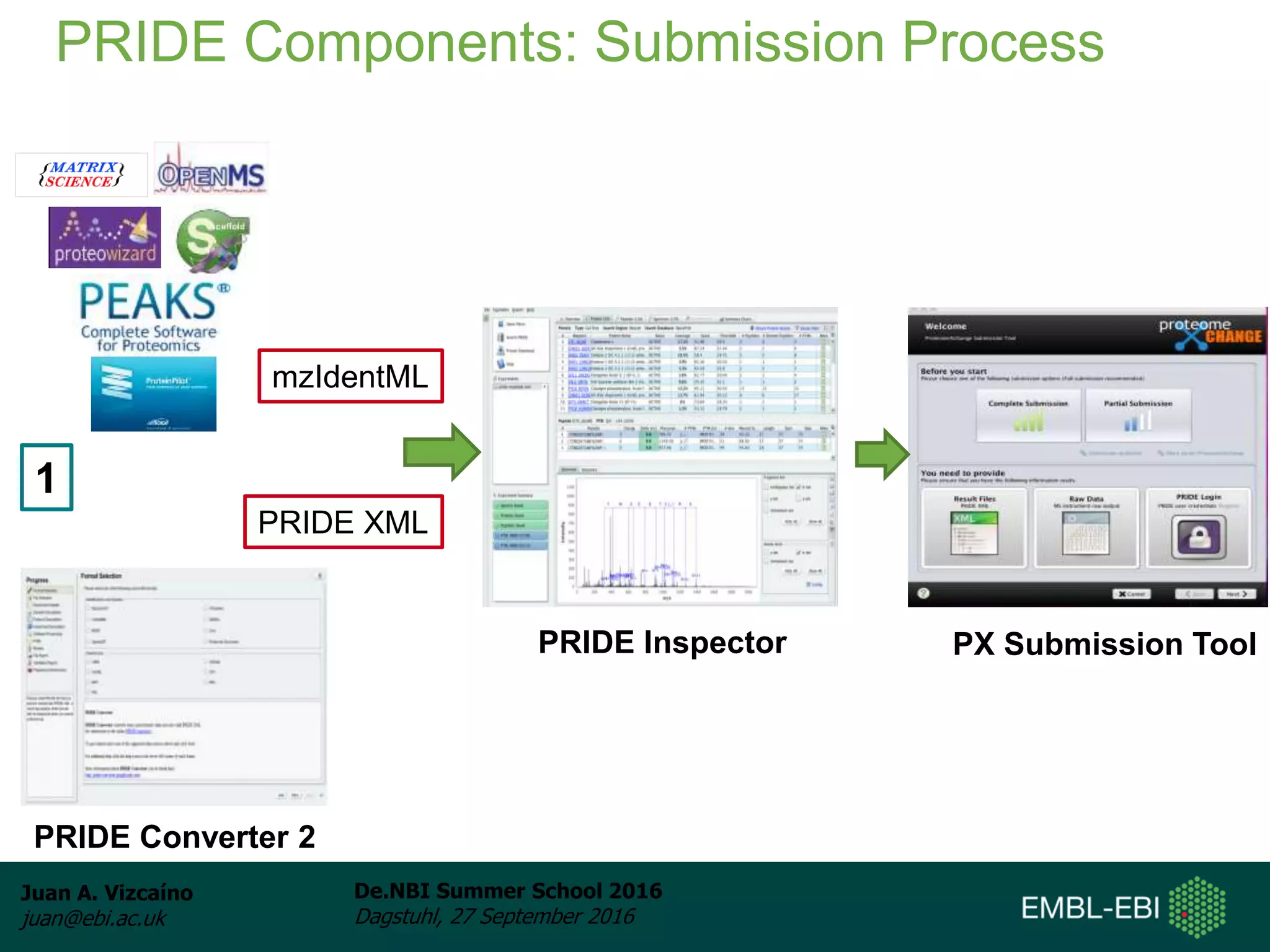
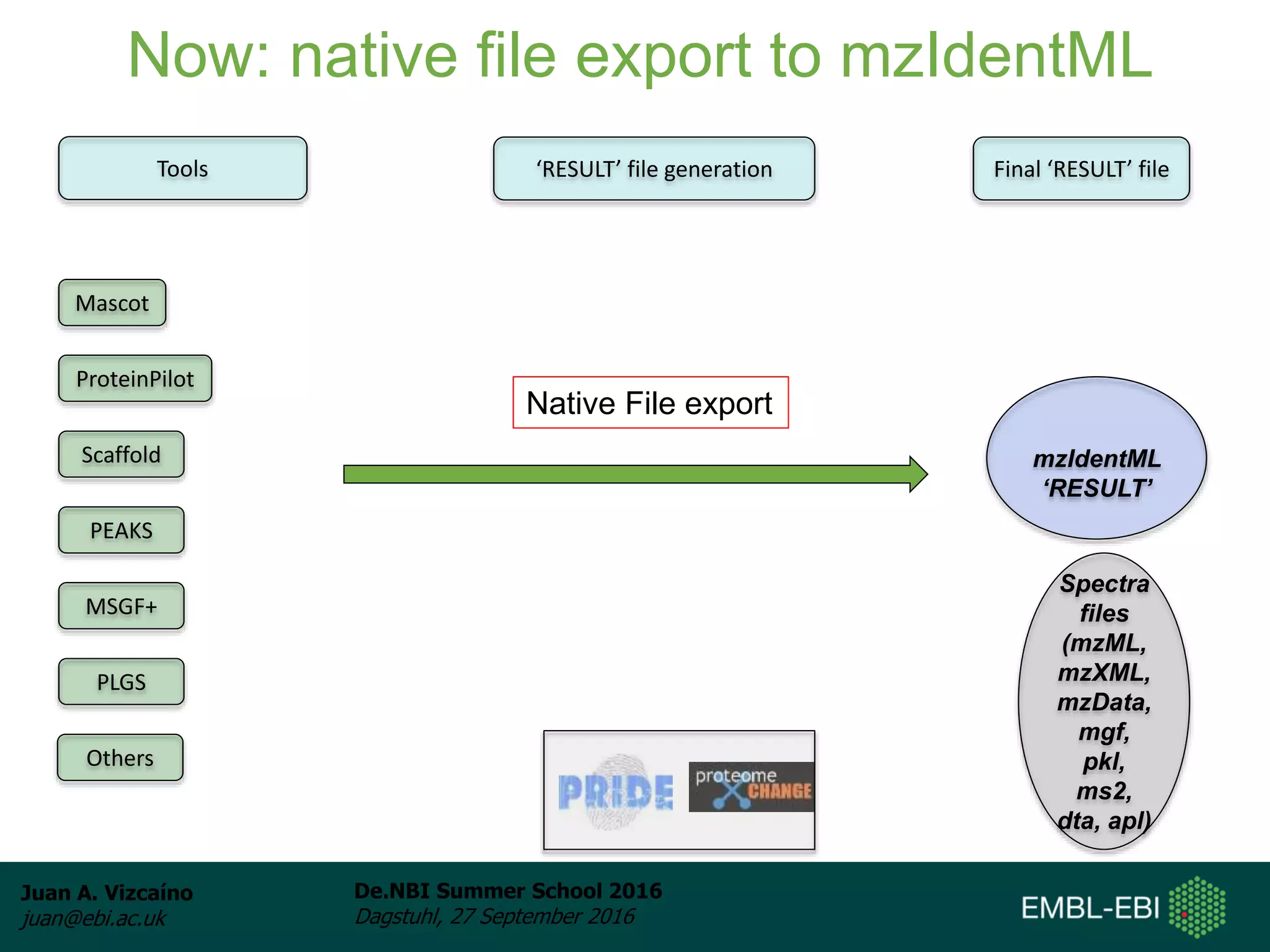
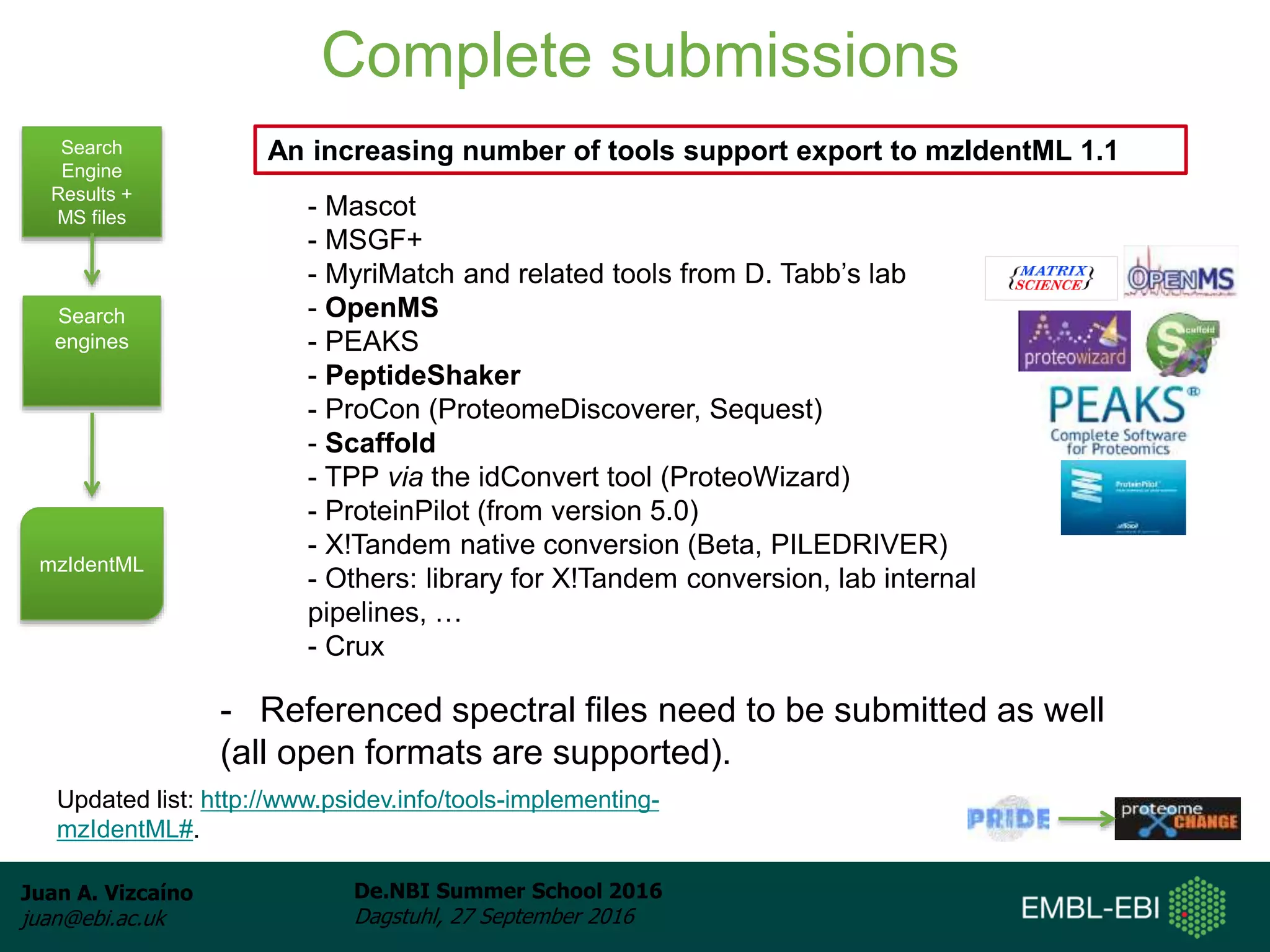
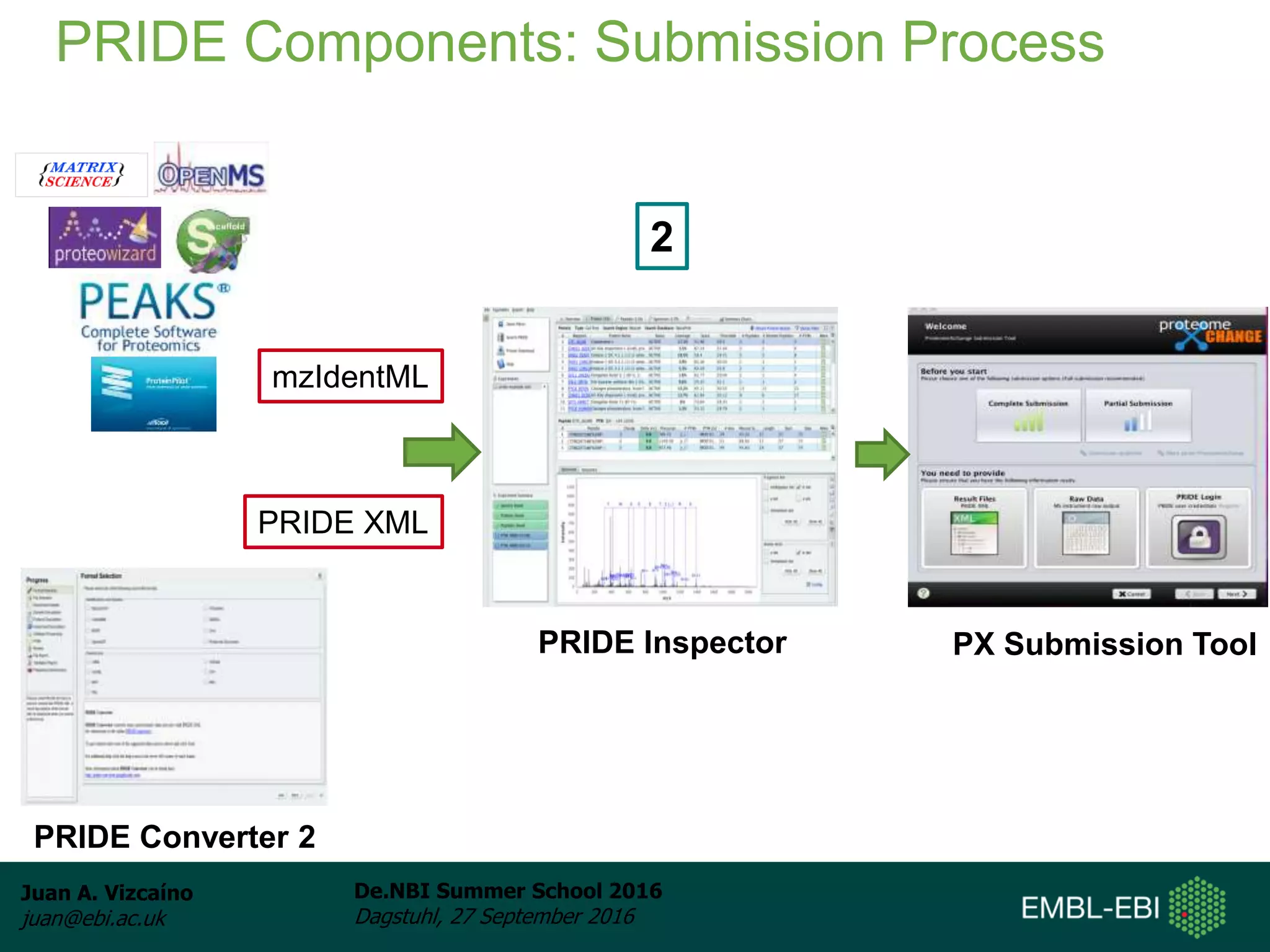
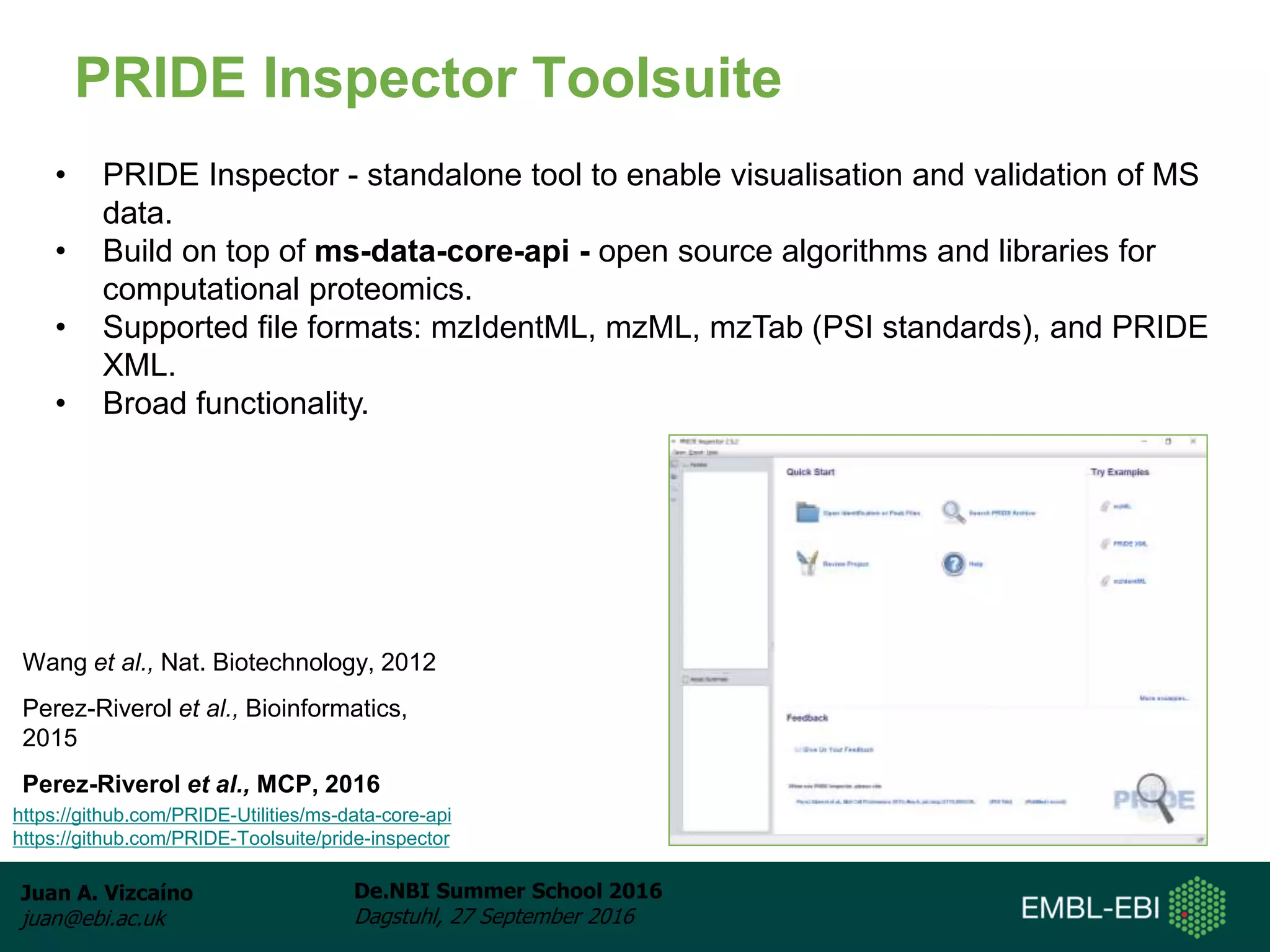
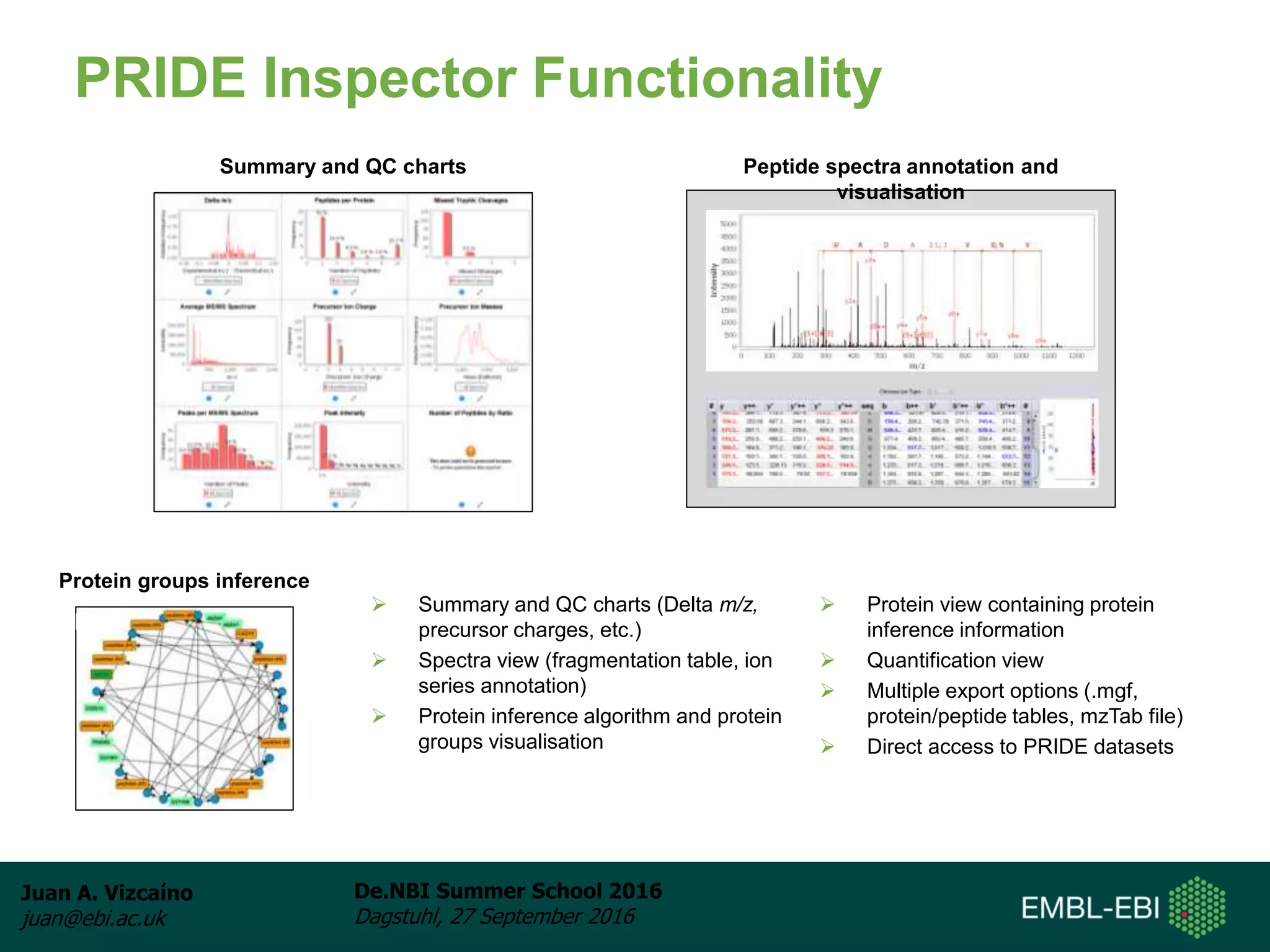
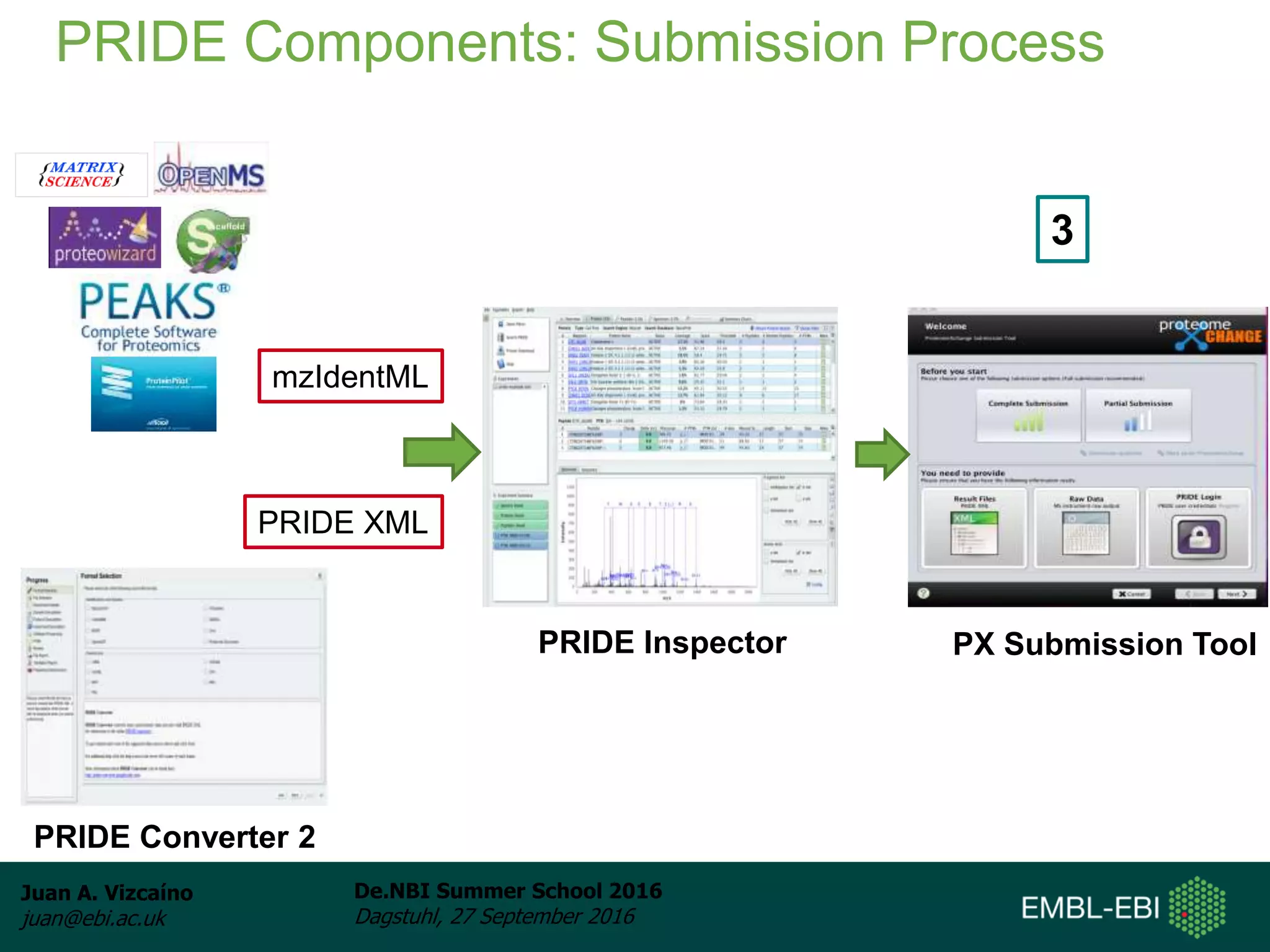
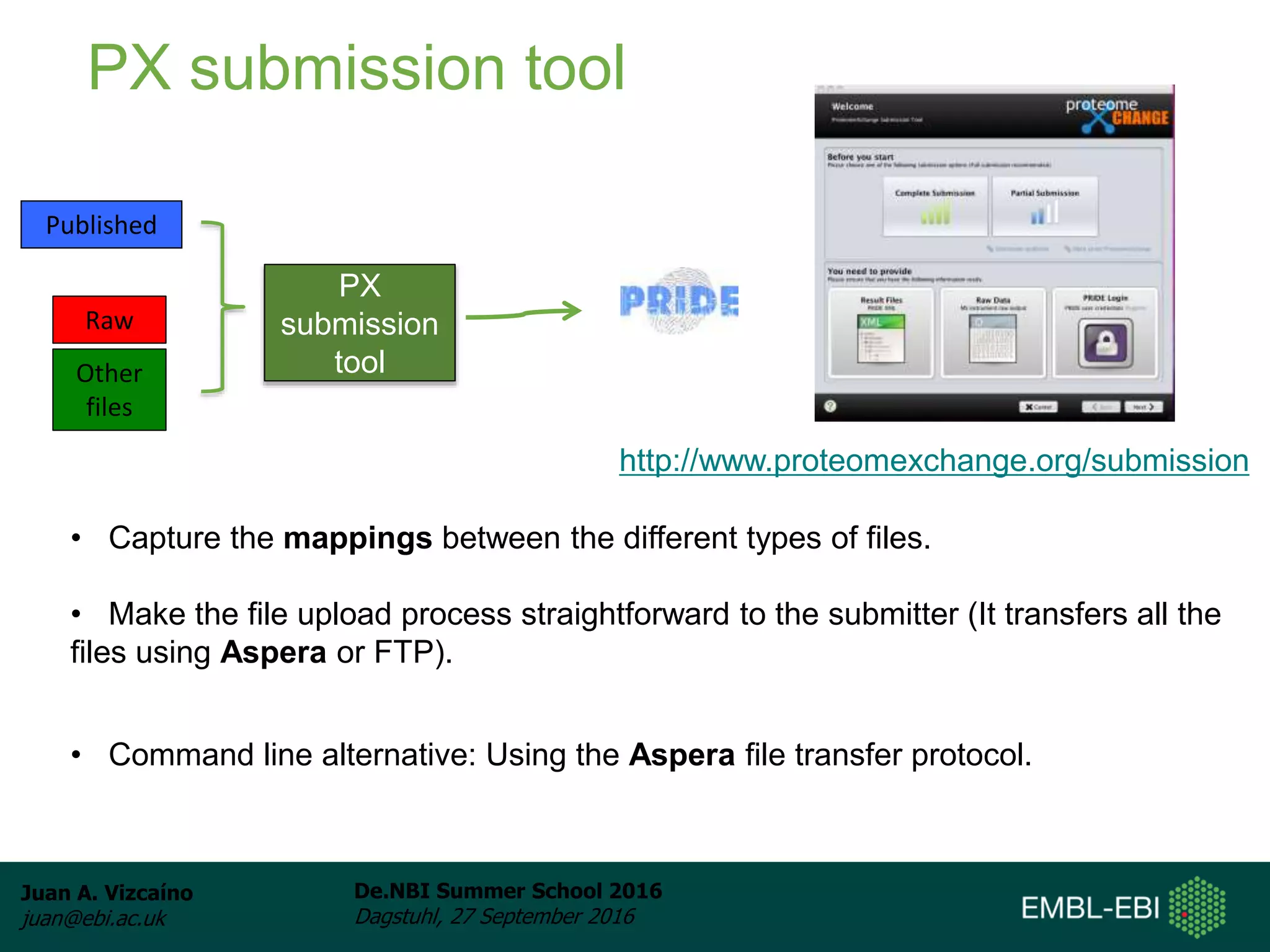
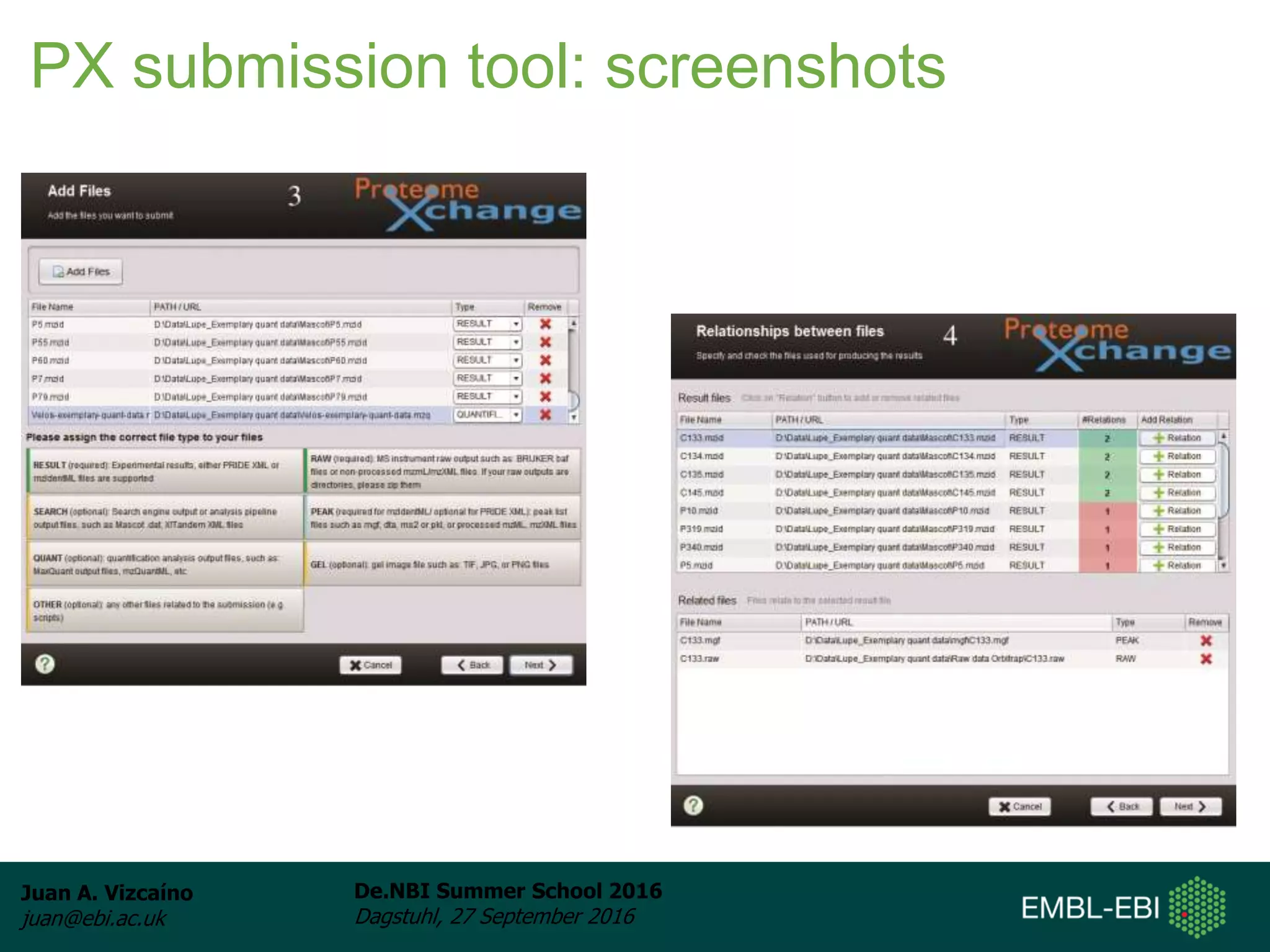
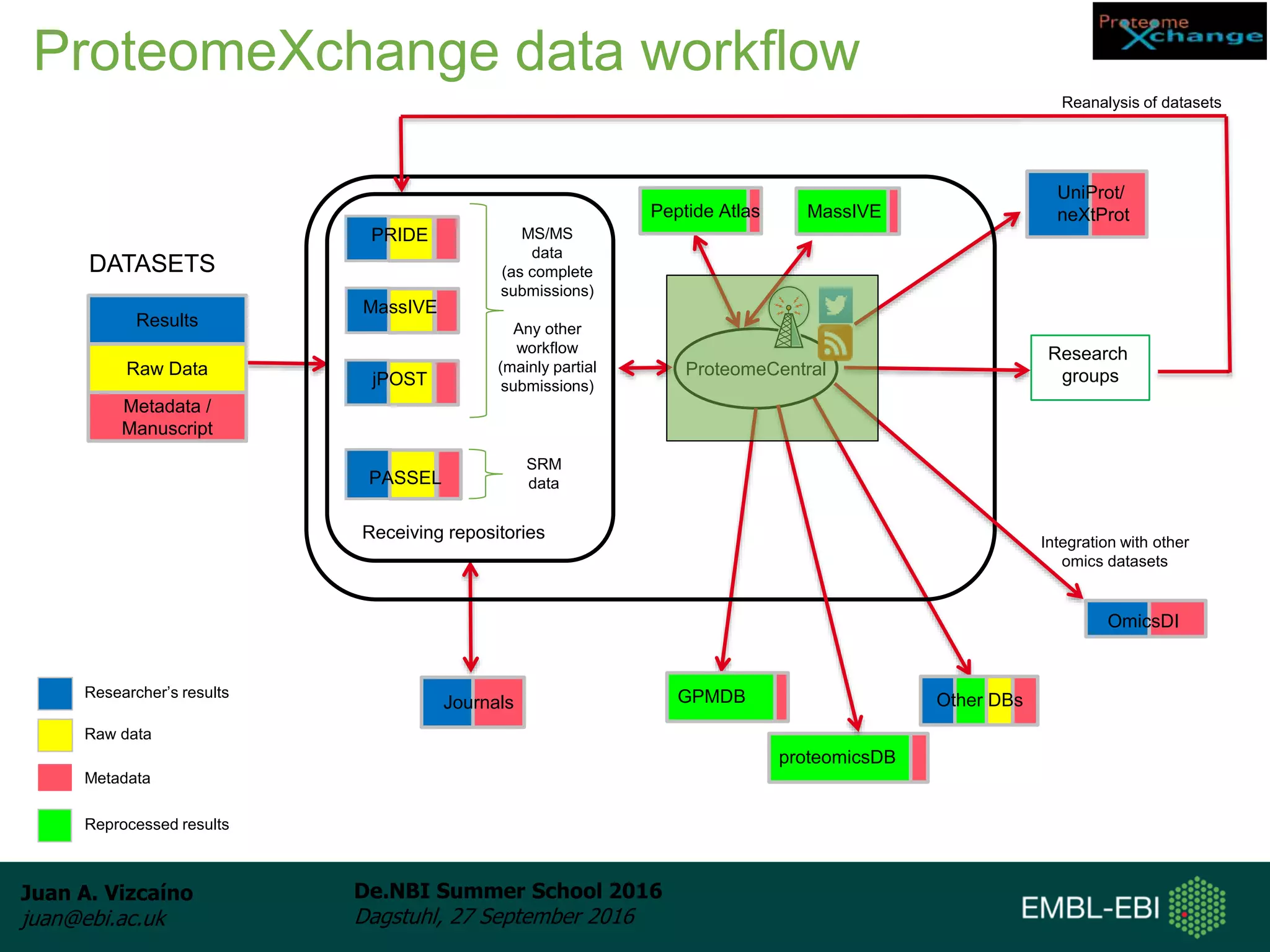
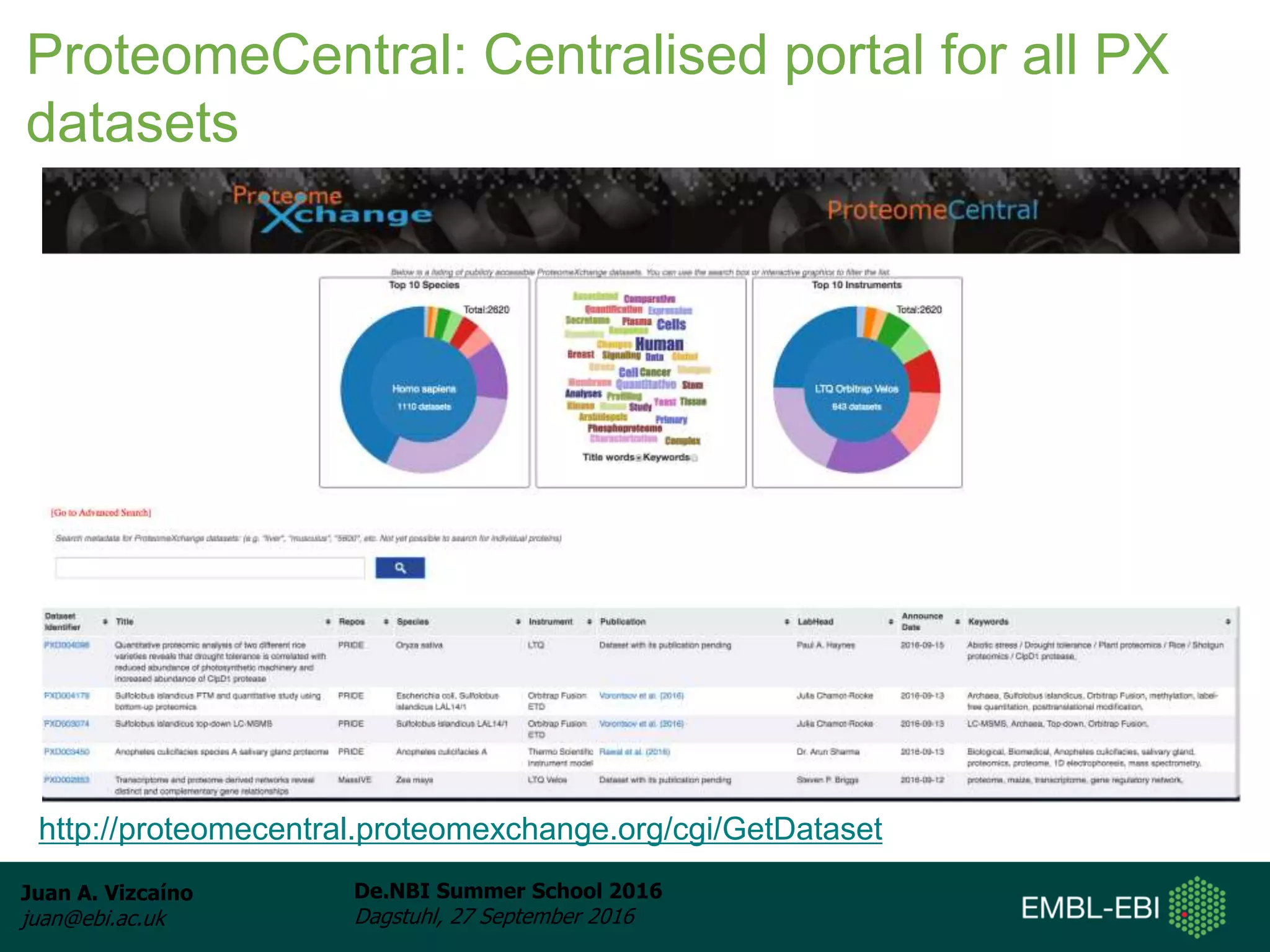
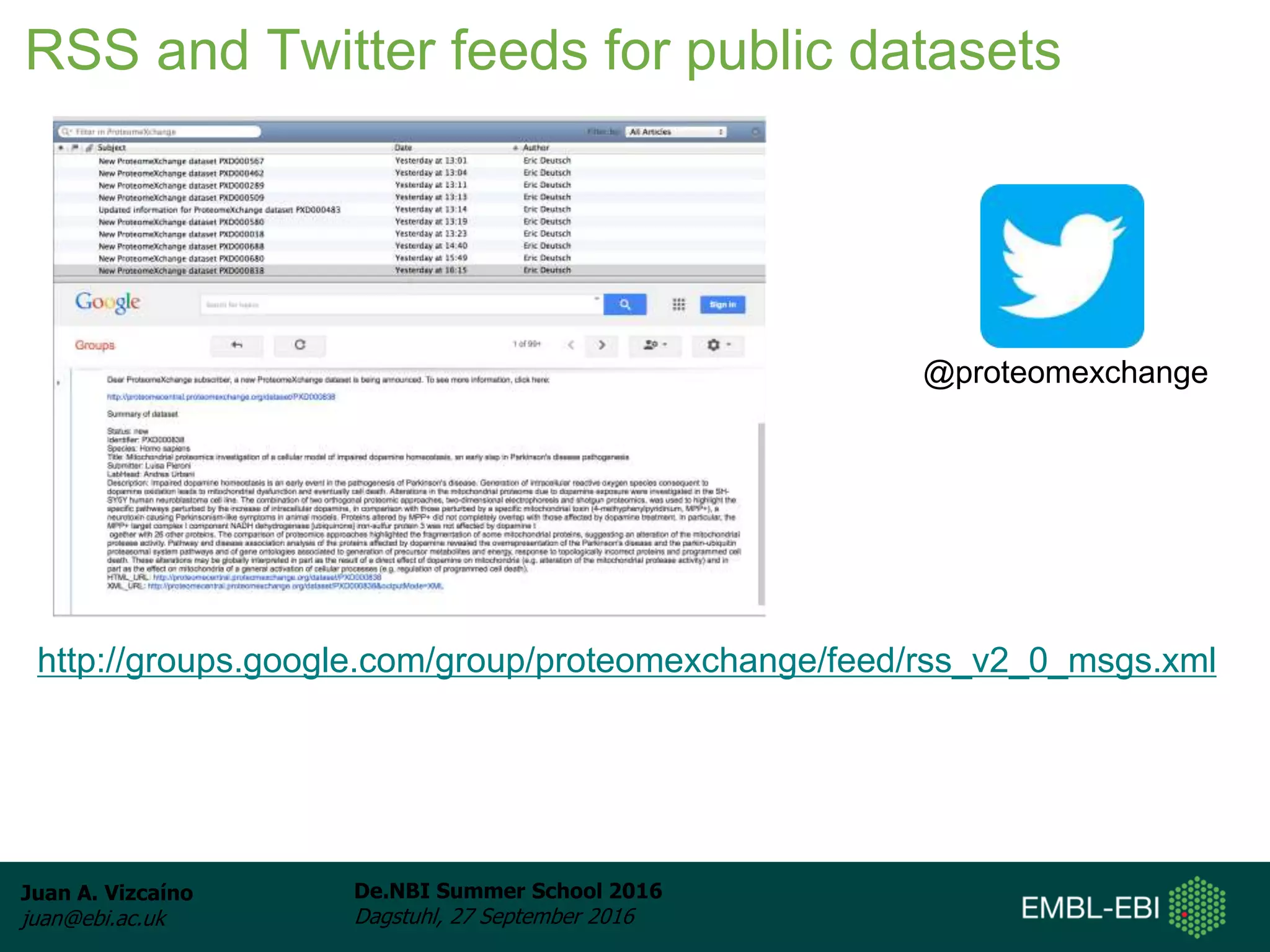
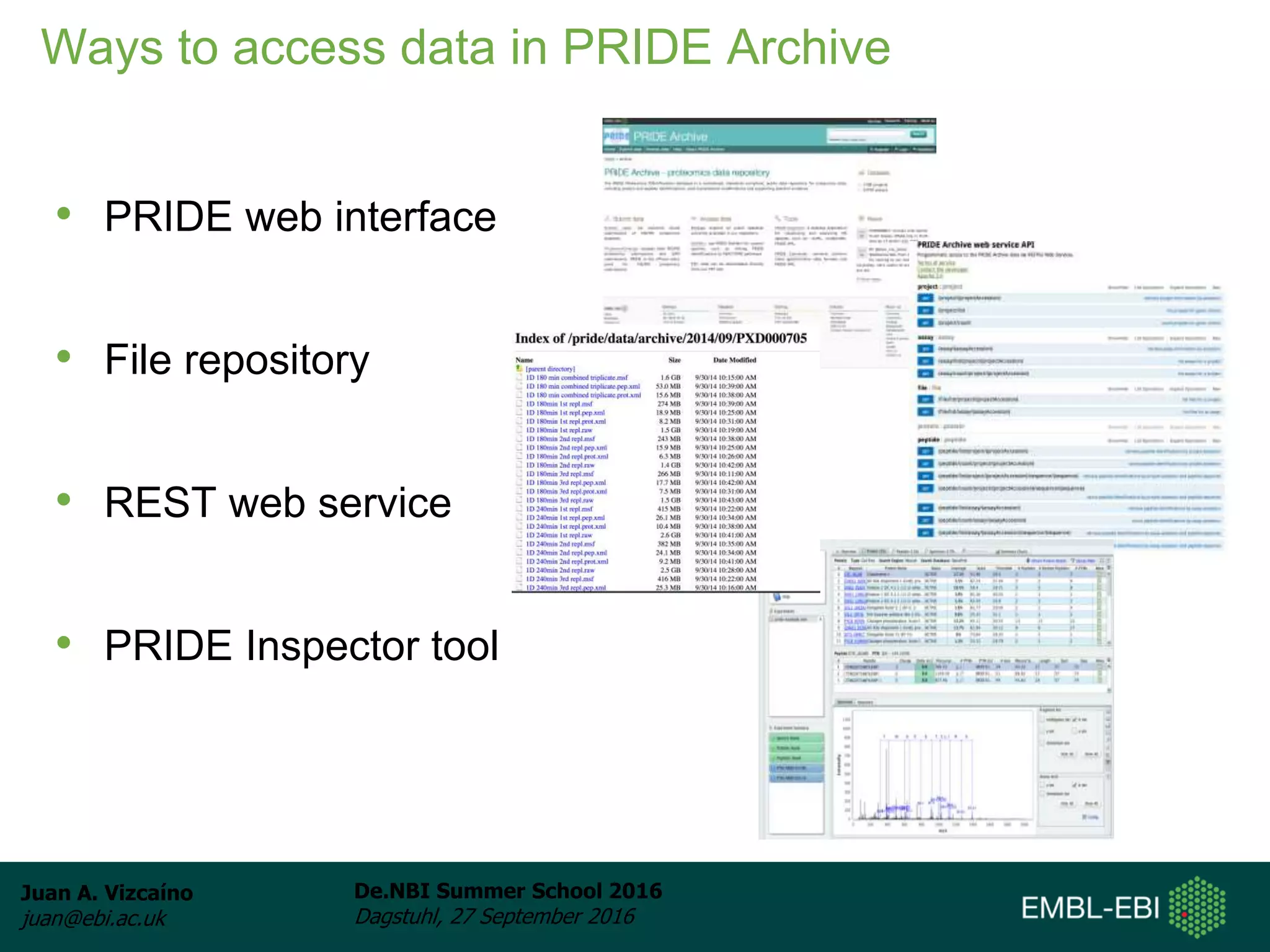
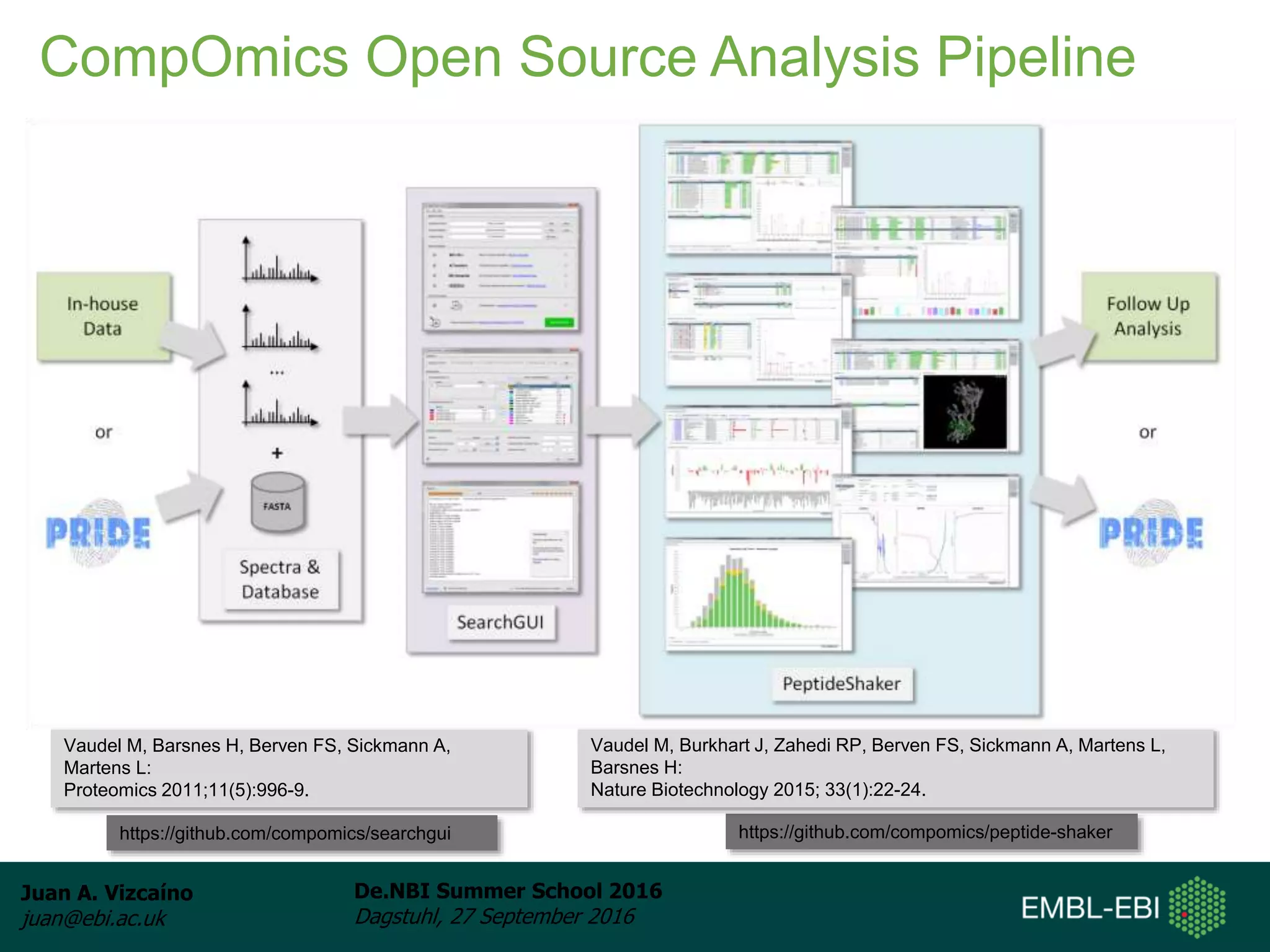
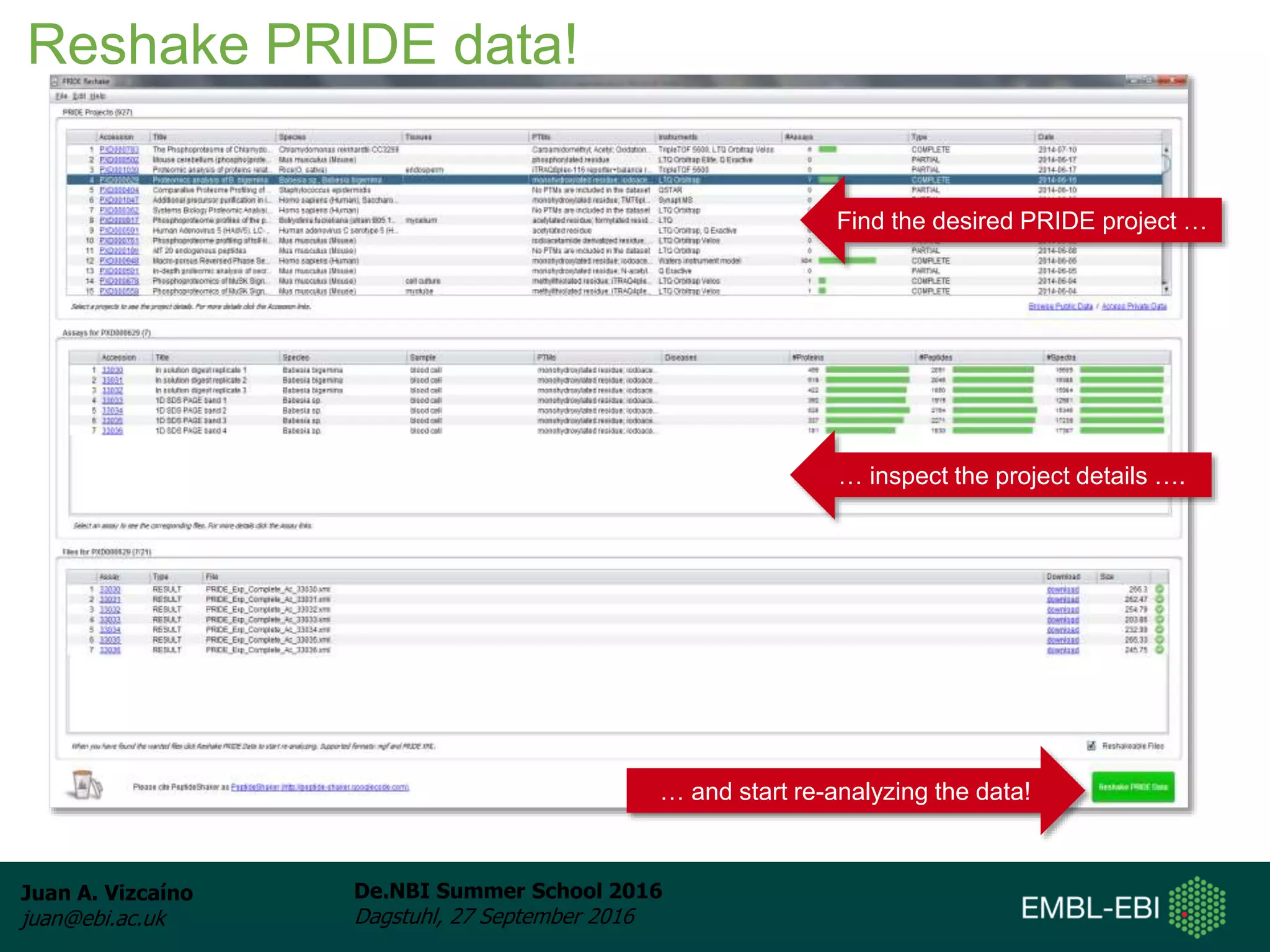
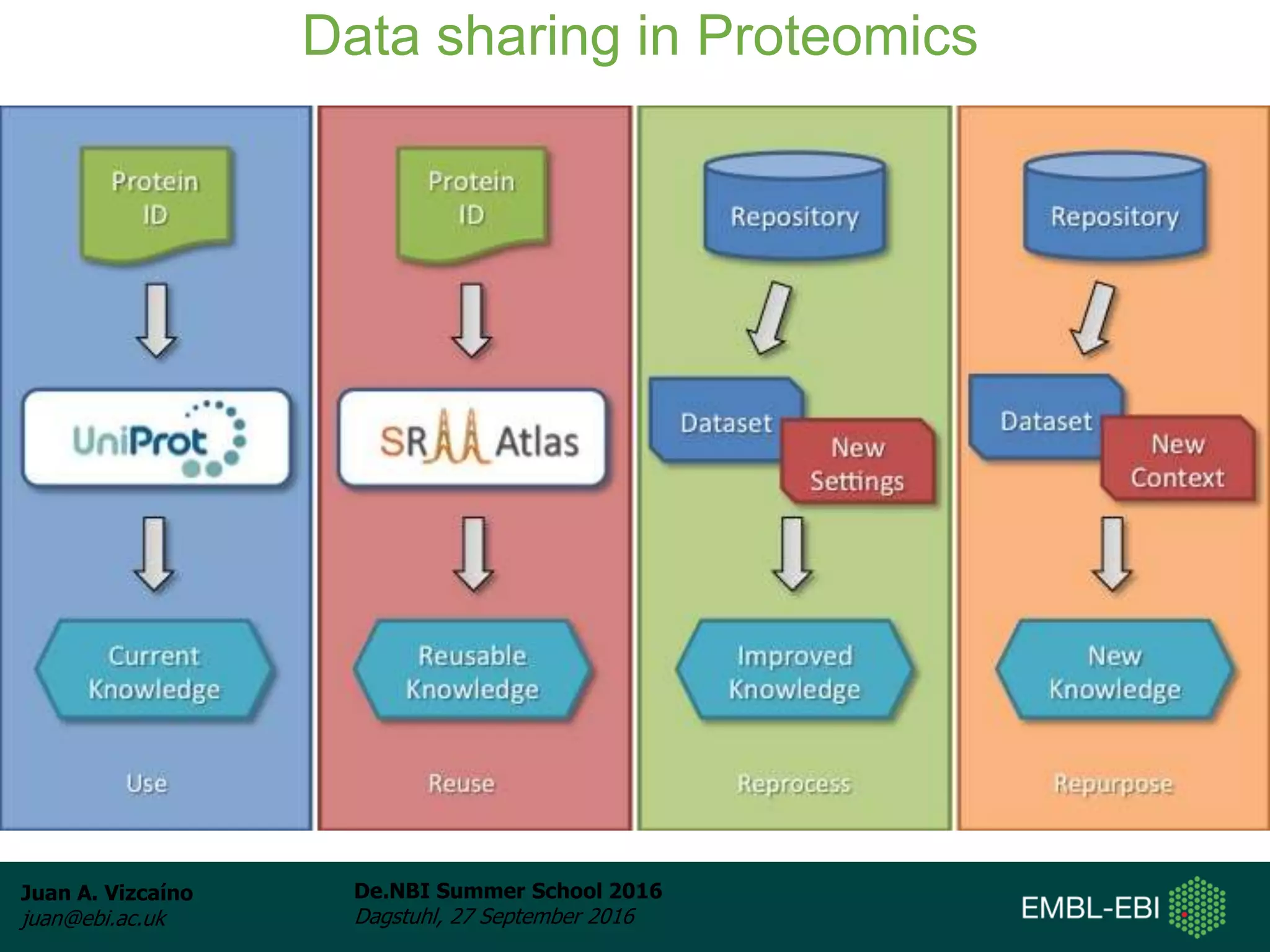
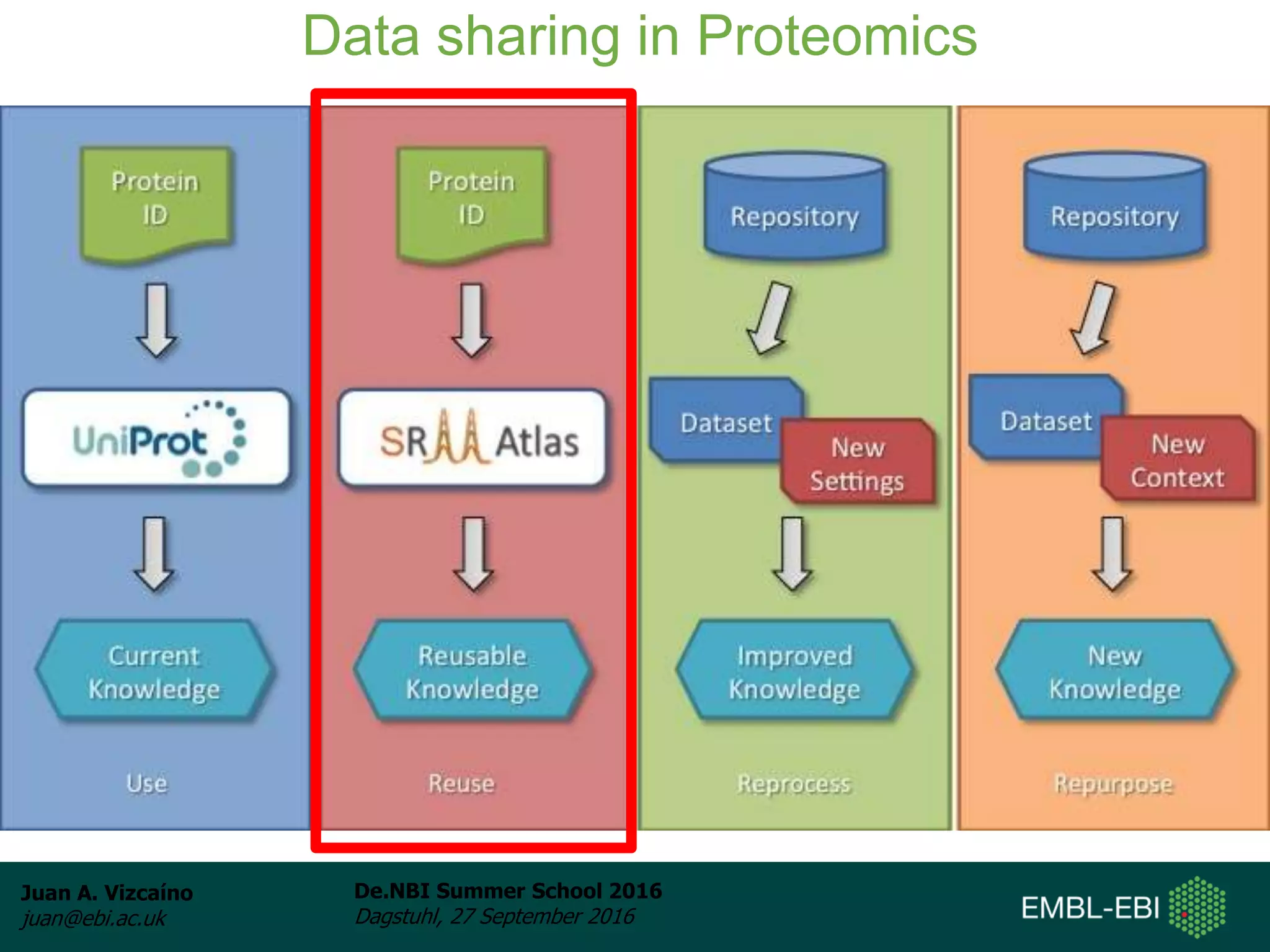
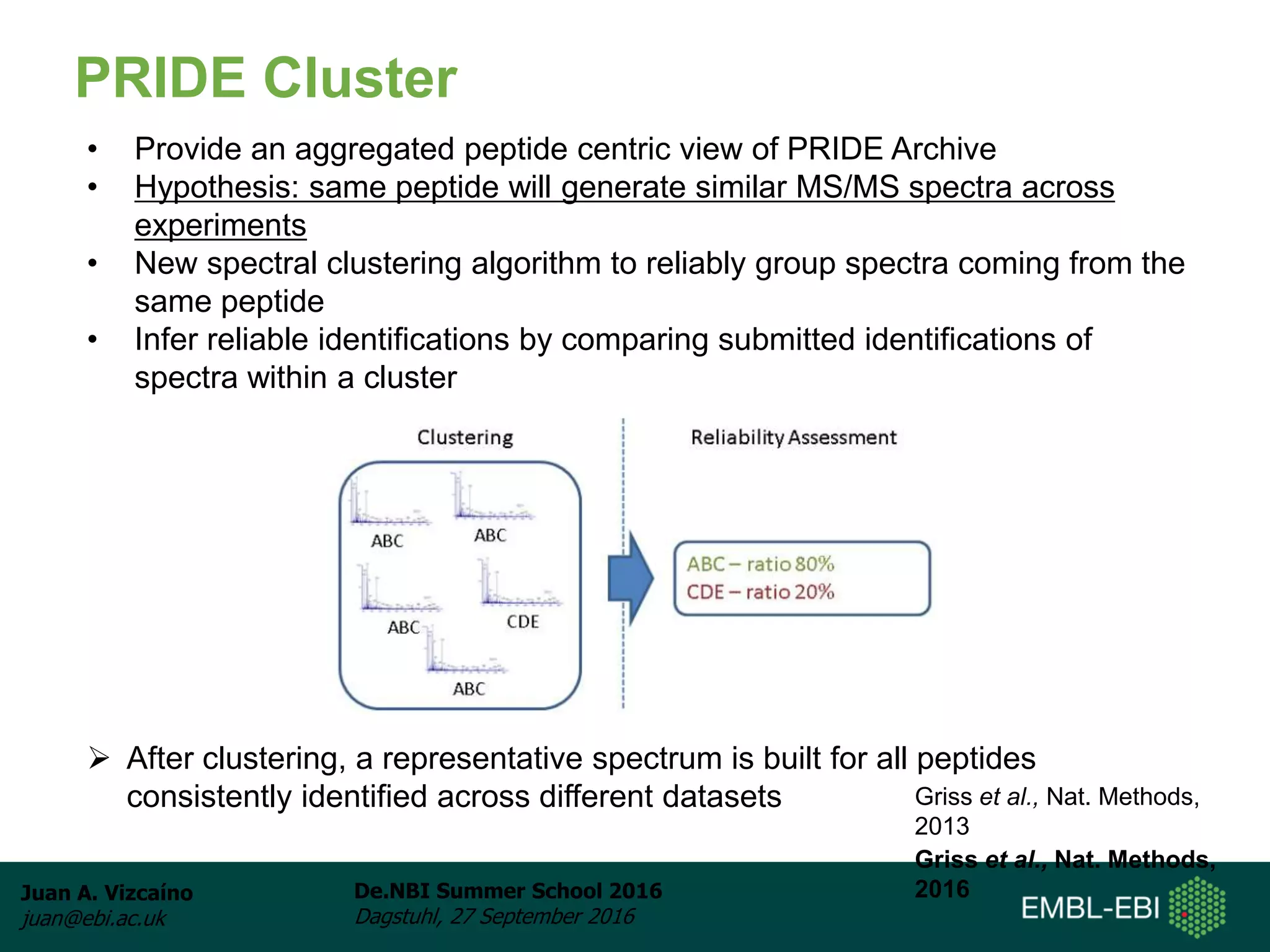
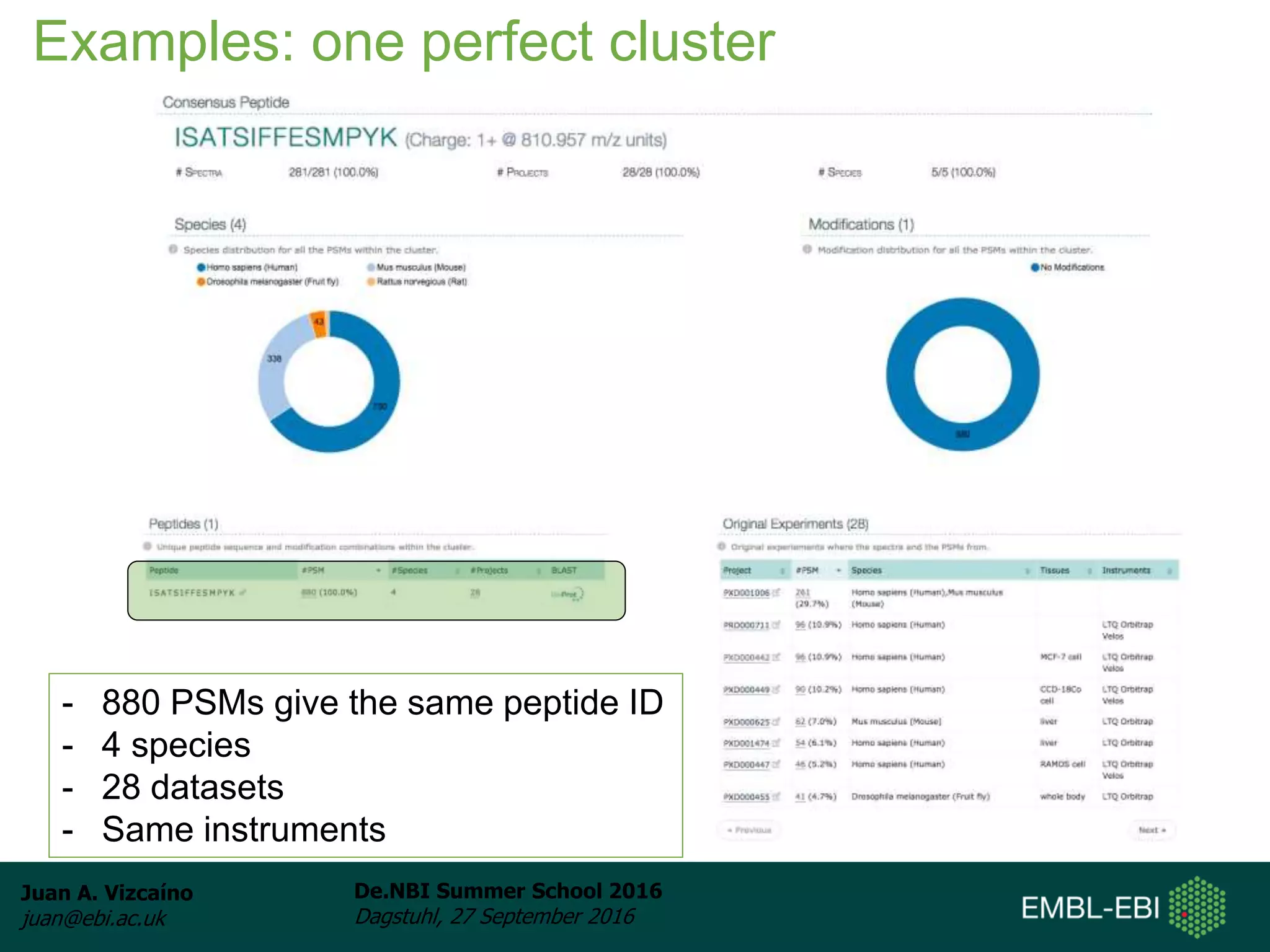
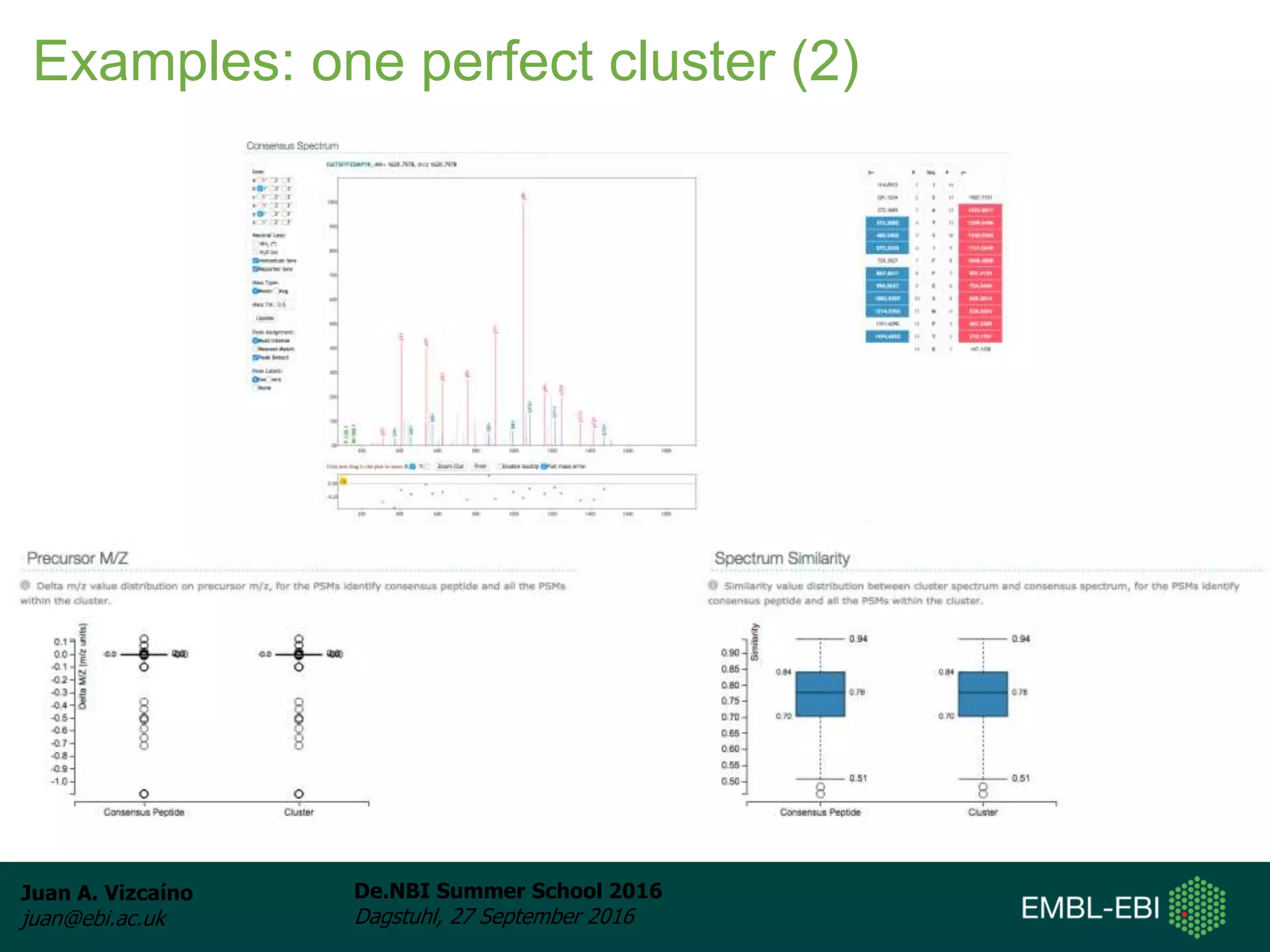
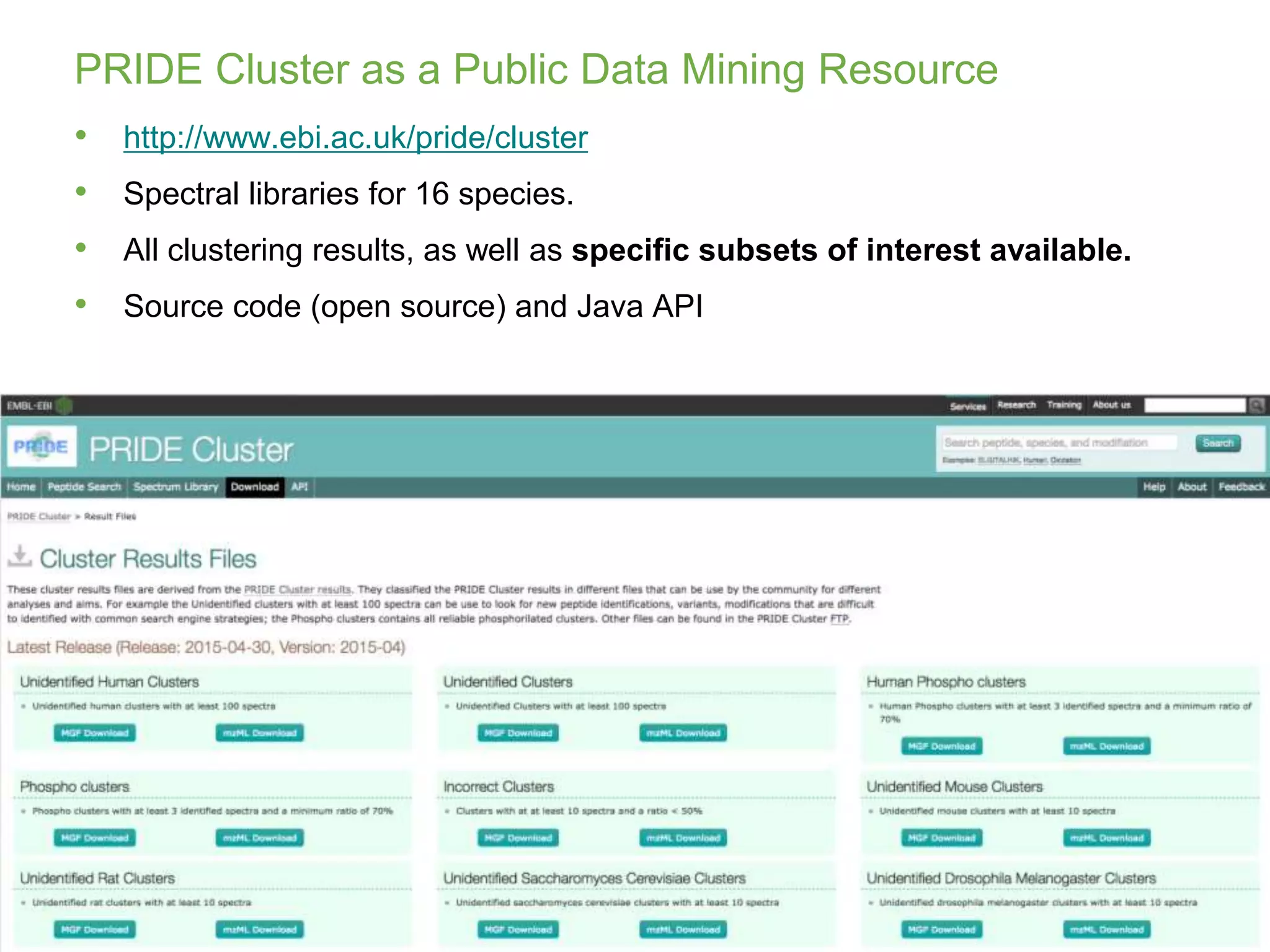
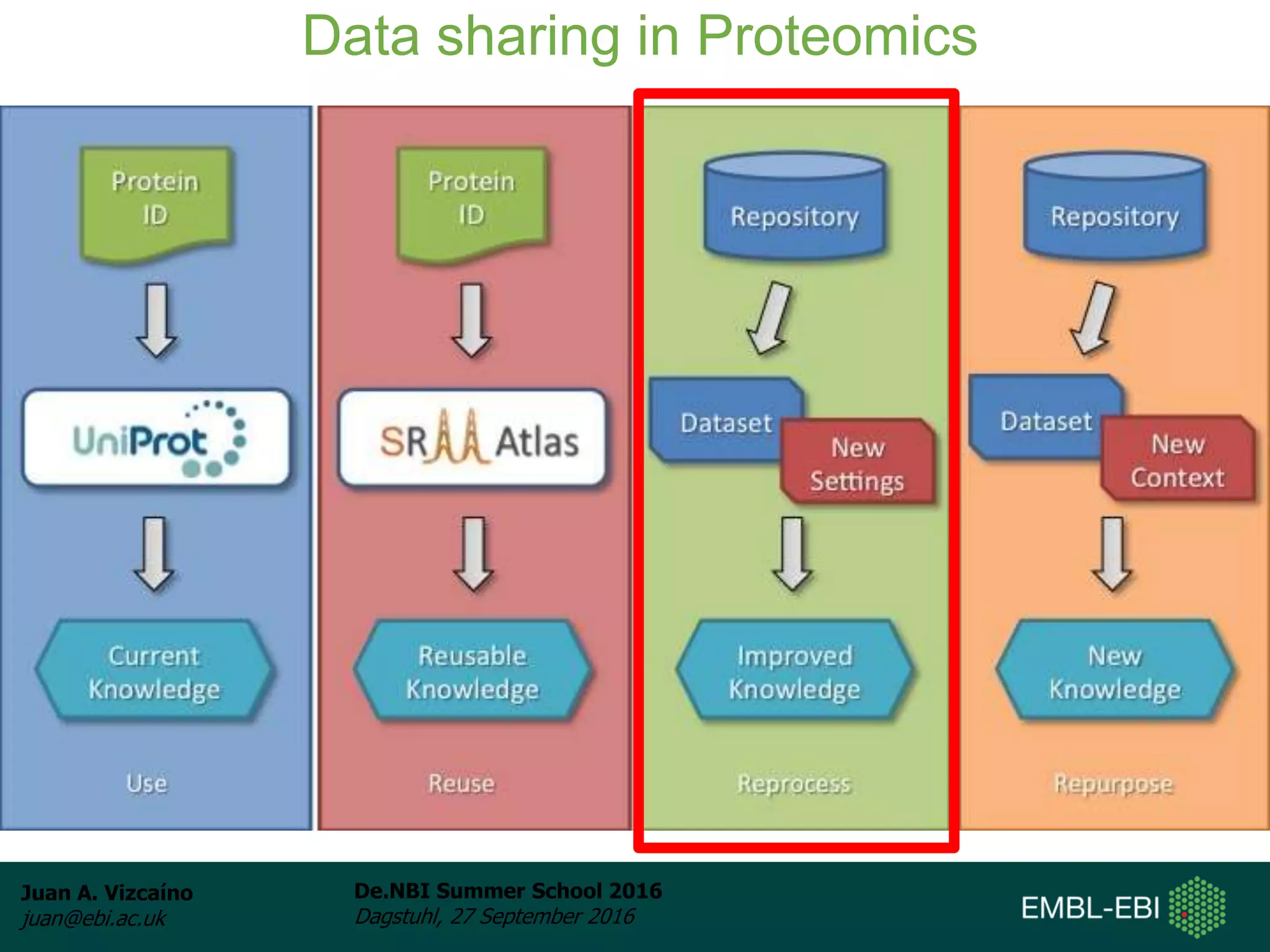
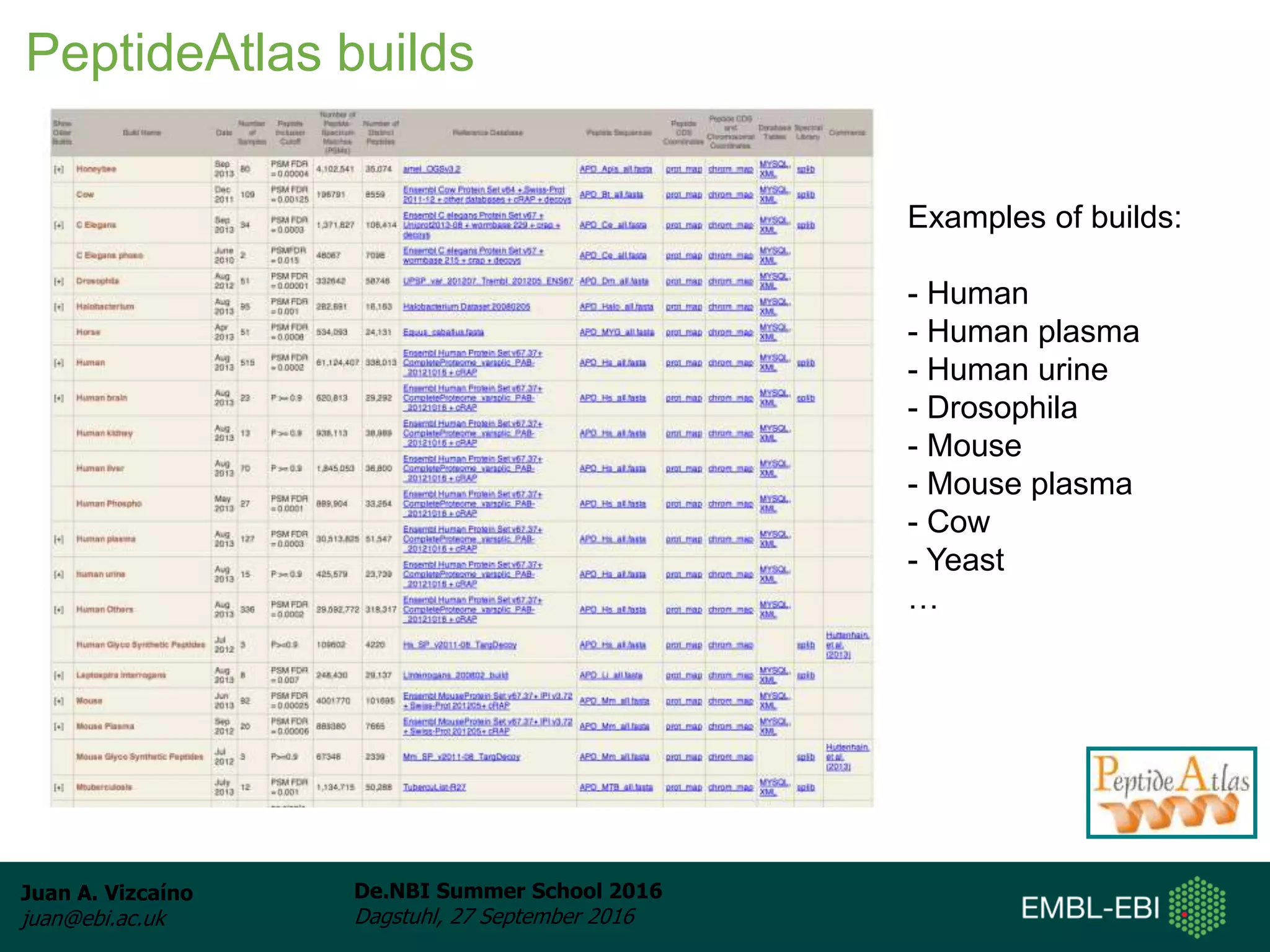
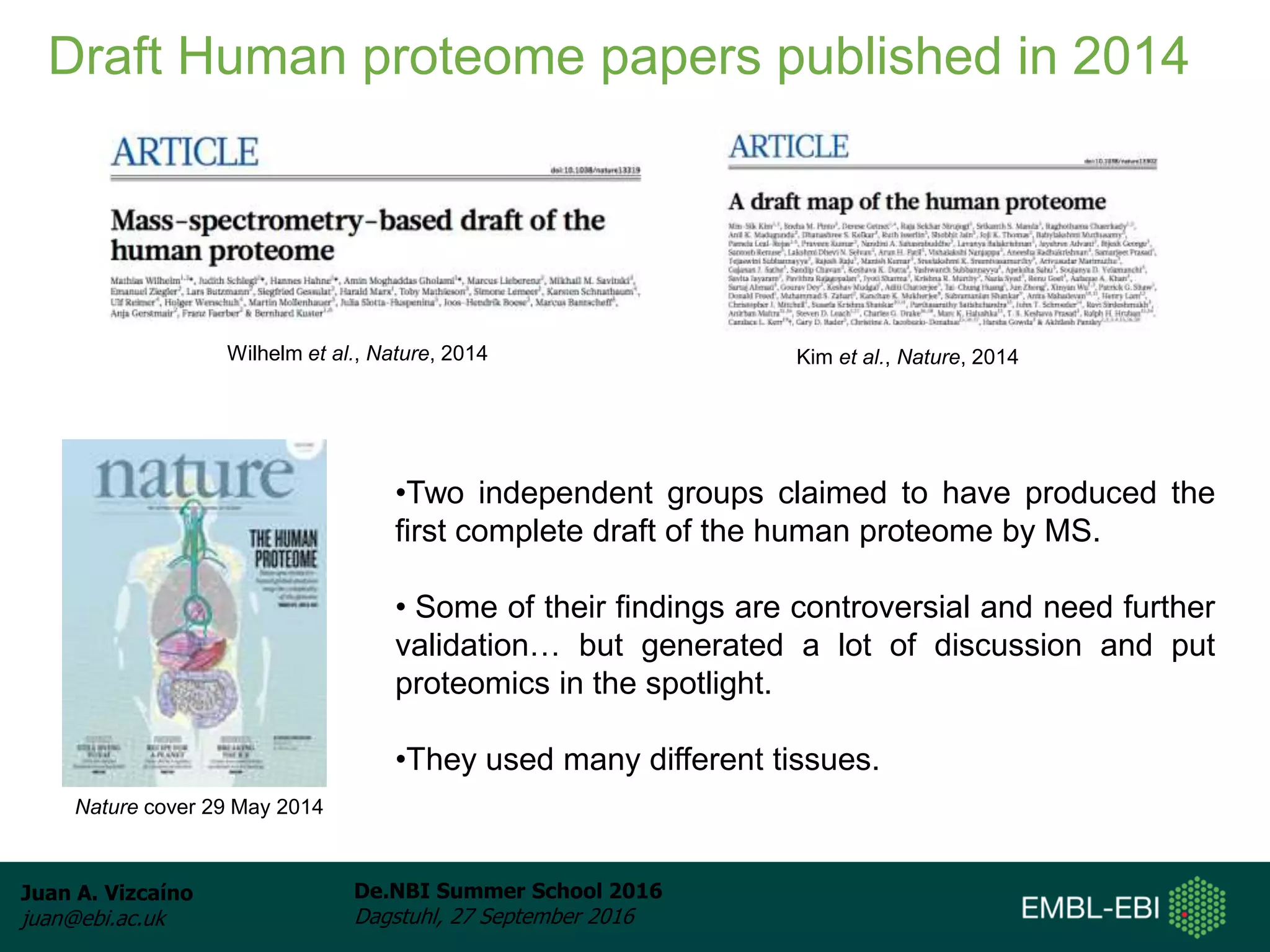
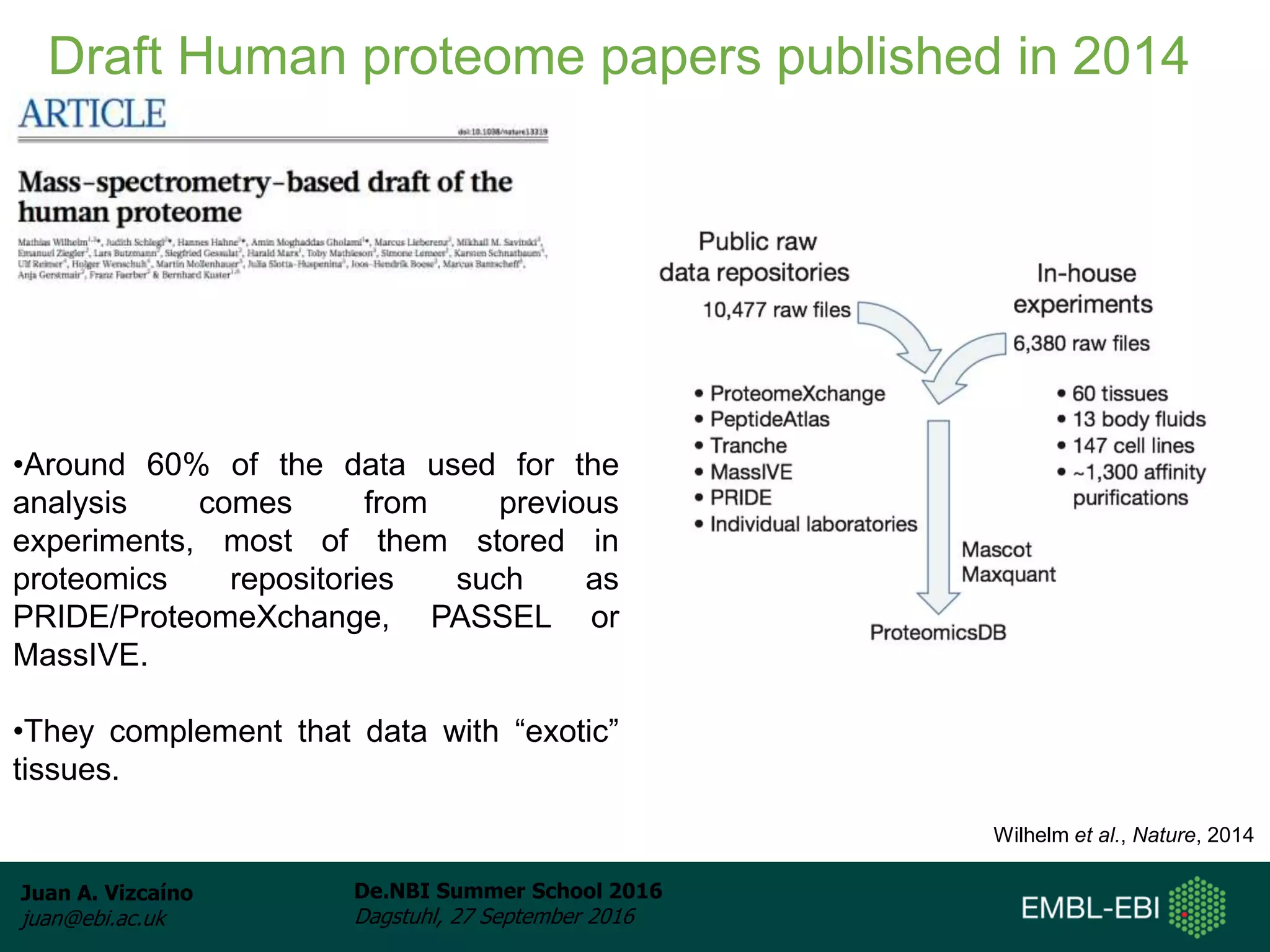
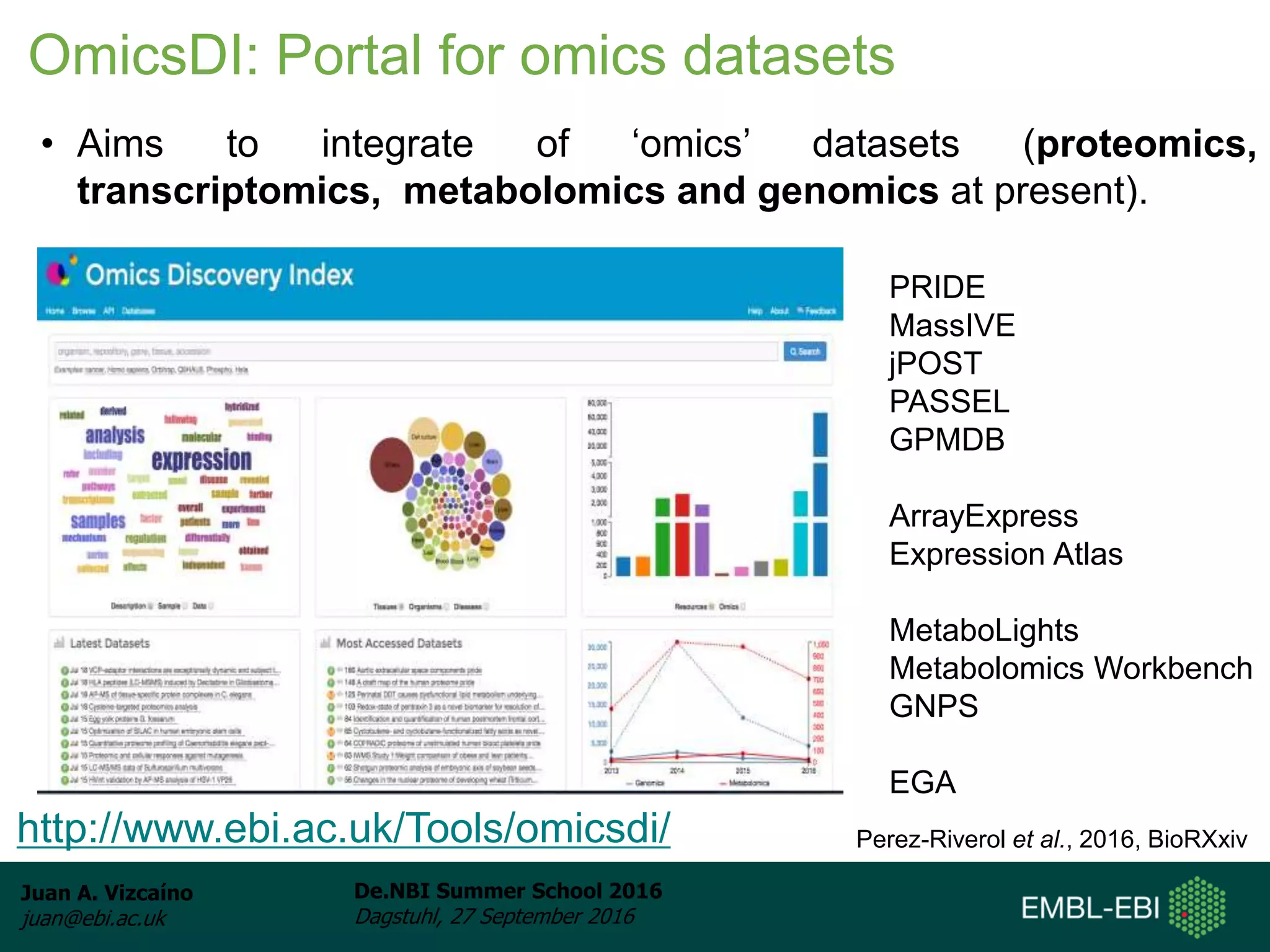
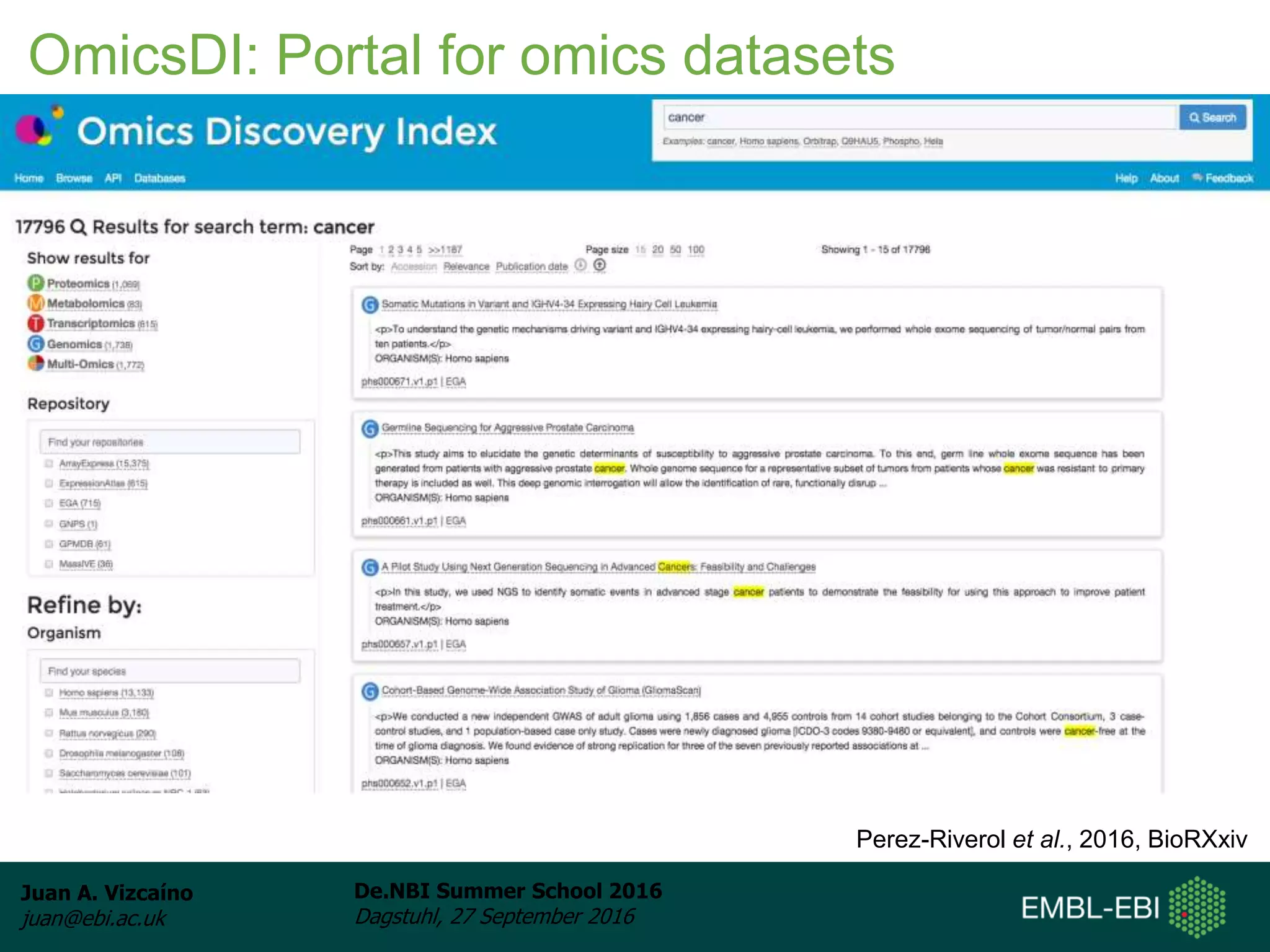
The document discusses PRIDE and ProteomeXchange, which are resources that support the deposition of proteomics data to public repositories. PRIDE stores mass spectrometry-based proteomics data, and is one of the repositories that is part of ProteomeXchange, a framework that allows standard submission of proteomics data between major repositories. The document outlines the cultural change in proteomics towards public data sharing, and provides information on submitting proteomics data to PRIDE and accessing data deposited in PRIDE and ProteomeXchange.