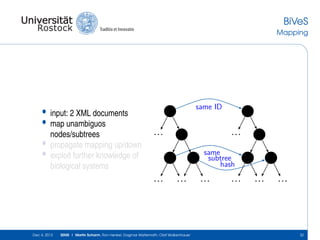
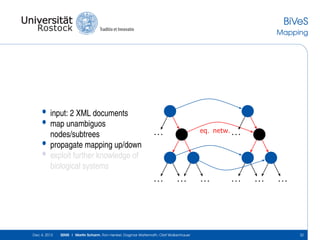
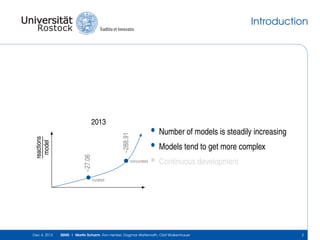
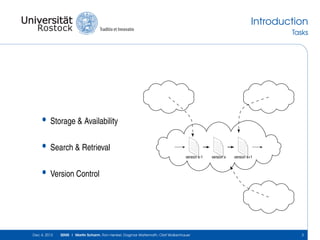
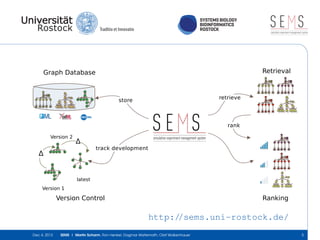
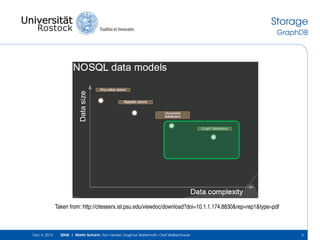
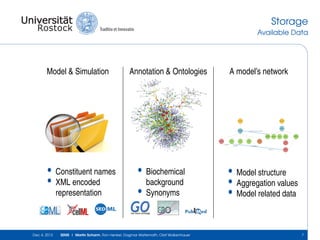
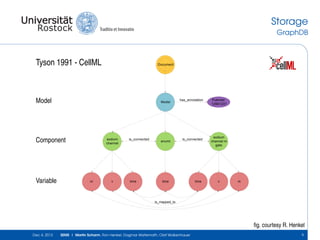
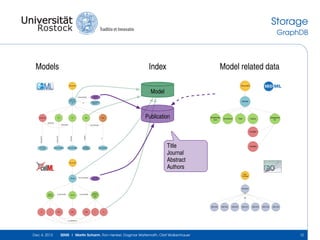
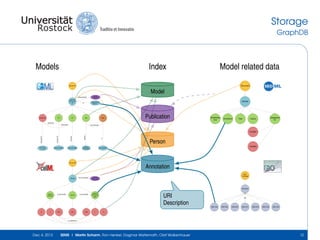
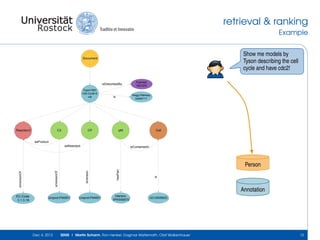
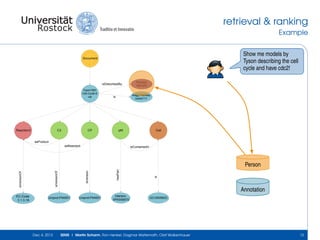
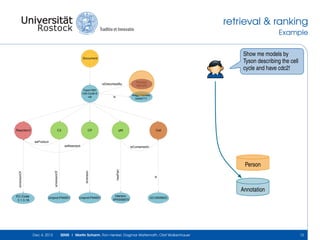
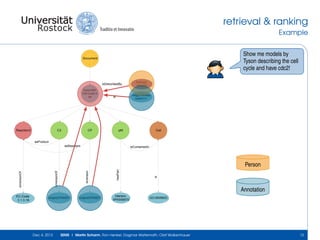
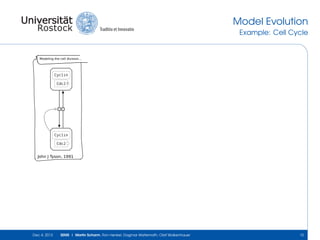
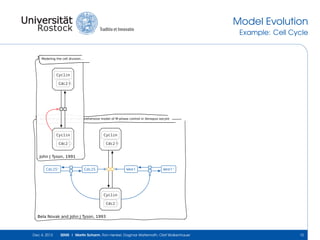
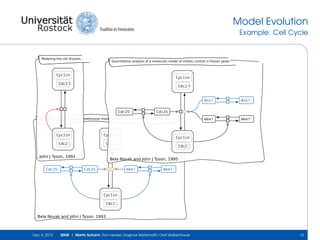
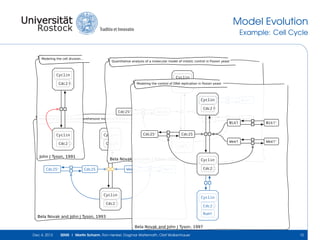
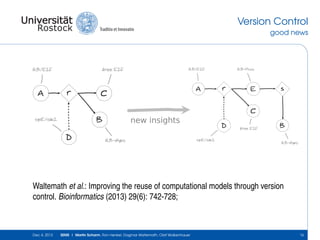
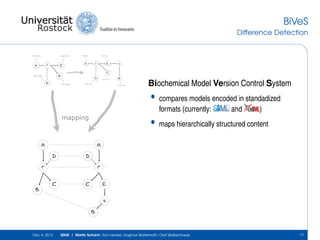
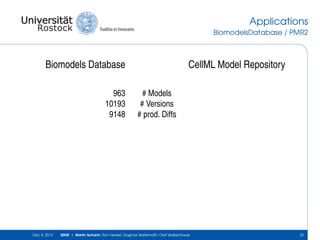
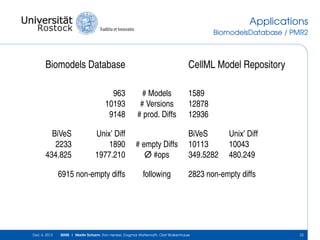
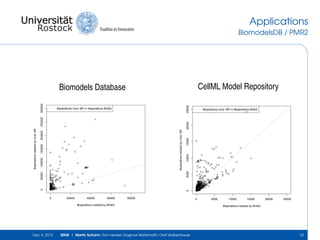
The document discusses the Simulation Experiment Management System (SEMS) developed for the management of computational models in systems biology and bioinformatics. It emphasizes the increasing complexity and quantity of models, highlighting essential tasks such as storage, retrieval, and version control. Various strategies and projects are being developed to support the community in managing these computational models efficiently.
























![Applications
Communicating Changes
Fixed the model → How to communicate the changes?
BiVeS Diff?
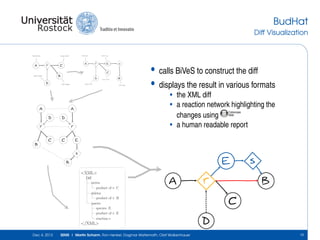
<?xml version="1.0" encoding="UTF-8" standalone="no"?>
<bives type="fullDiff">
<update/>
<delete>
<node id="1" oldChildNo="1" oldParent="/listOfReactions[1]/reaction[
<attribute id="2" name="rdf:resource" oldPath="/listOfReactions[1]/r
<node id="5" oldChildNo="4" oldParent="/listOfReactions[1]/reaction[
<node id="6" oldChildNo="1" oldParent="/listOfReactions[1]/reaction[
<attribute id="7" name="species" oldPath="/listOfReactions[1]/reacti
</delete>
<insert>
<attribute id="3" name="metaid" newPath="/listOfReactions[1]/reactio
<attribute id="4" name="metaid" newPath="/listOfReactions[1]/reactio
<attribute id="9" name="metaid" newPath="/listOfReactions[1]/reactio
<node id="10" newChildNo="1" newParent="/listOfReactions[1]/reaction
much smarter, but still 24 lines..
Dec 4, 2013
SEMS | Martin Scharm, Ron Henkel, Dagmar Waltemath, Olaf Wolkenhauer
21](https://image.slidesharecdn.com/martin-scharm-ebi-dec2013-131219111047-phpapp02/85/Improving-the-Management-of-Computational-Models-Invited-talk-at-the-EBI-48-320.jpg)





![BiVeS
Integration
jvm
network
cmd
import de.unirostock.sems.bives.api.SBMLDiff;
[...]
SBMLDiff differ = new SBMLDiff (sbmlFileA, sbmlFileB);
differ.mapTrees ();
String graph = differ.getCRNGraphML ();
[...]
Dec 4, 2013
SEMS | Martin Scharm, Ron Henkel, Dagmar Waltemath, Olaf Wolkenhauer
29](https://image.slidesharecdn.com/martin-scharm-ebi-dec2013-131219111047-phpapp02/85/Improving-the-Management-of-Computational-Models-Invited-talk-at-the-EBI-60-320.jpg)
![BiVeS
Integration
network
jvm
cmd
curl -d ’{
"get":
[
"documentType",
"xmlDiff"
],
"files":
{
"versionA":"http://your.db/path/to/versionA.sbml",
"versionB":"http://your.db/path/to/versionA.sbml"
}
}’ http://bives.server.tld
Dec 4, 2013
SEMS | Martin Scharm, Ron Henkel, Dagmar Waltemath, Olaf Wolkenhauer
29](https://image.slidesharecdn.com/martin-scharm-ebi-dec2013-131219111047-phpapp02/85/Improving-the-Management-of-Computational-Models-Invited-talk-at-the-EBI-61-320.jpg)