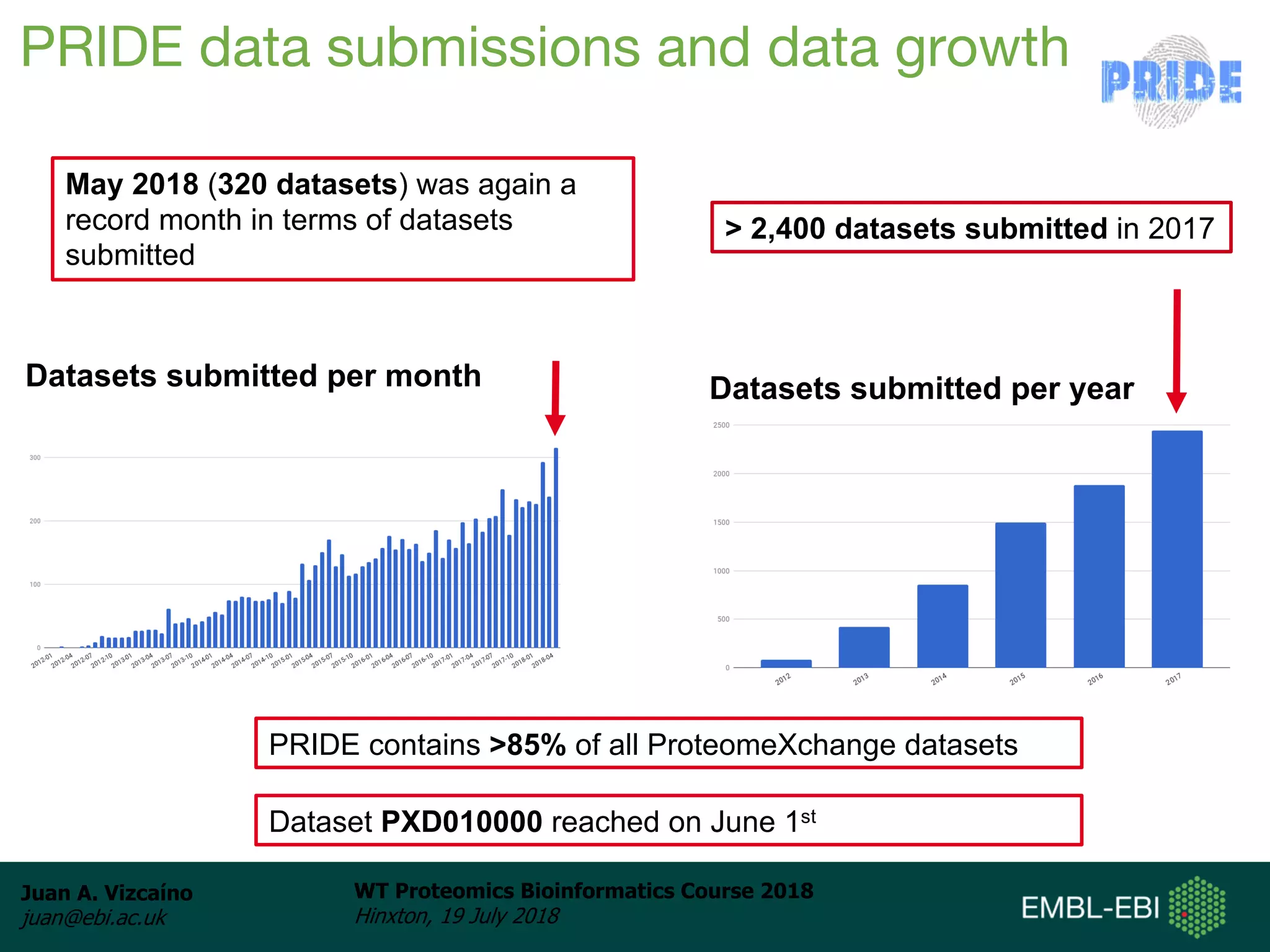
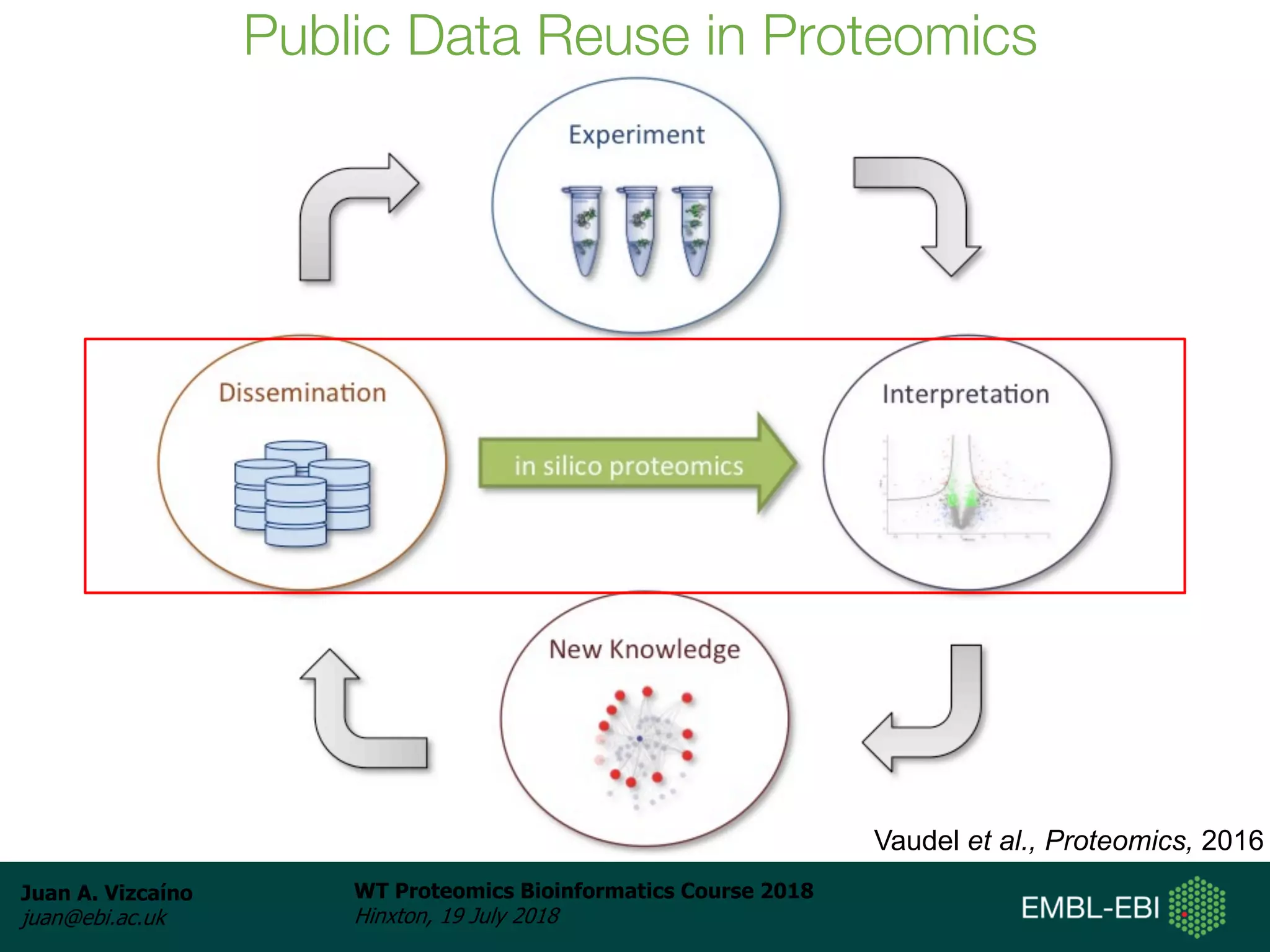
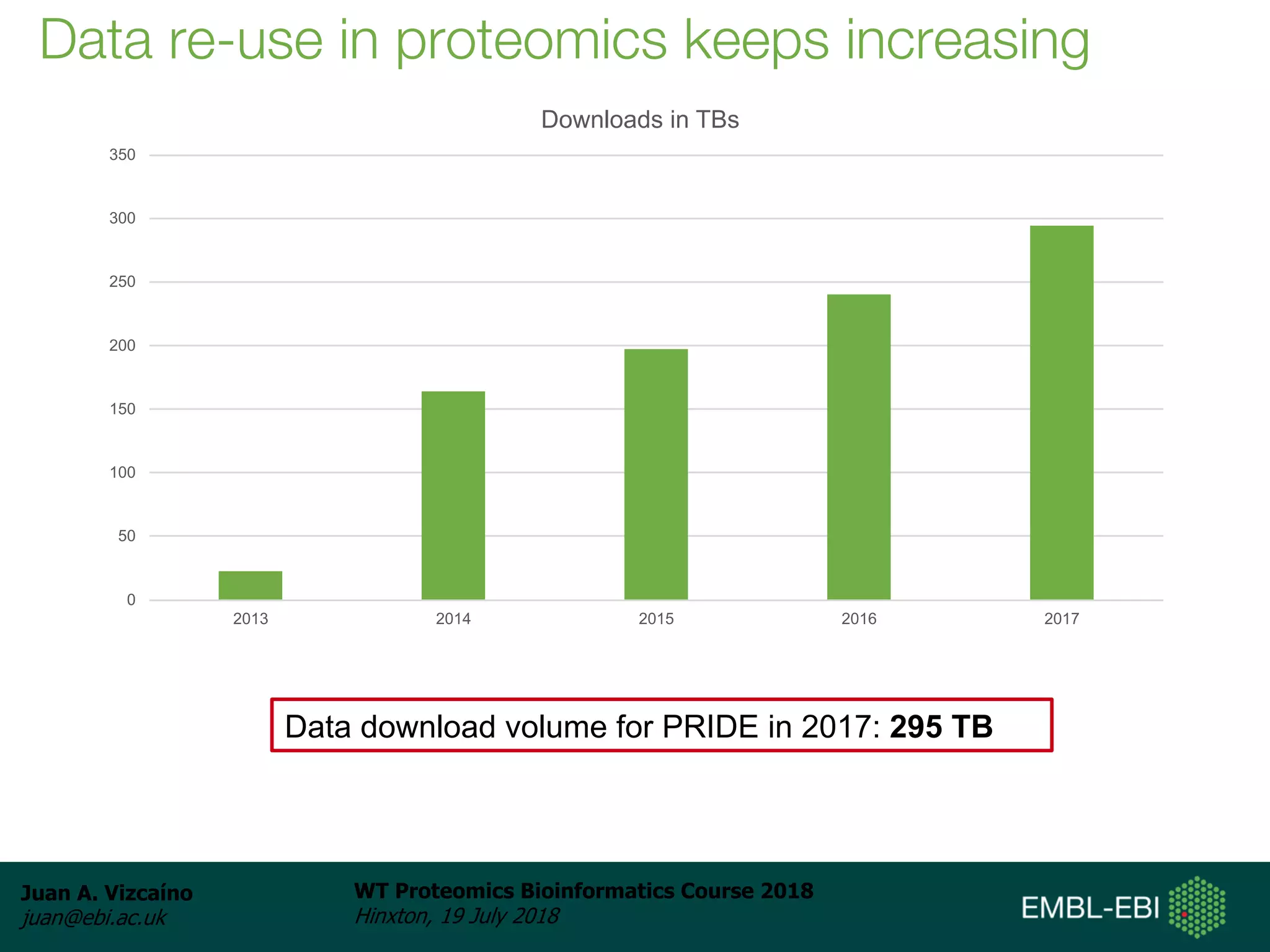
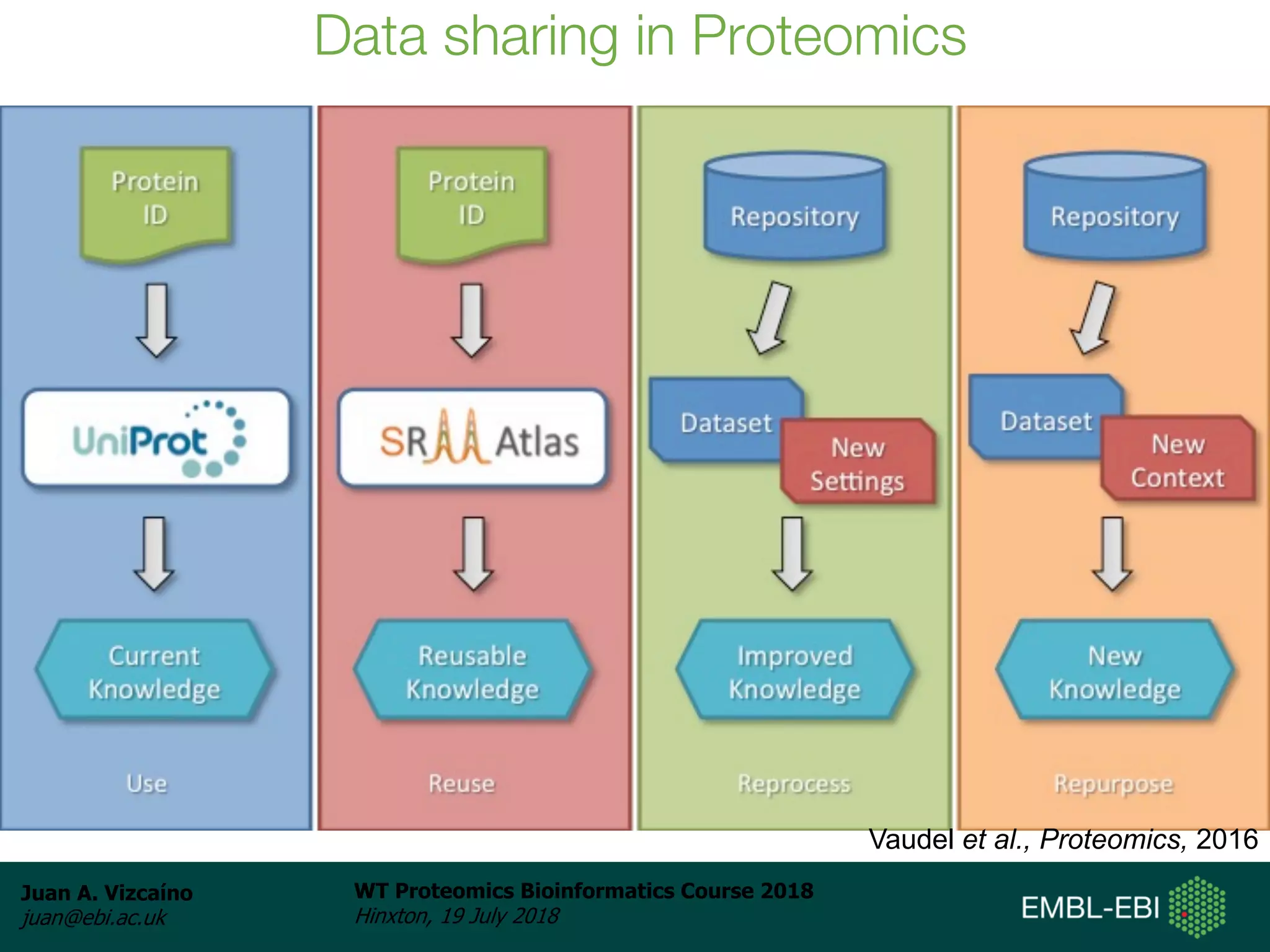
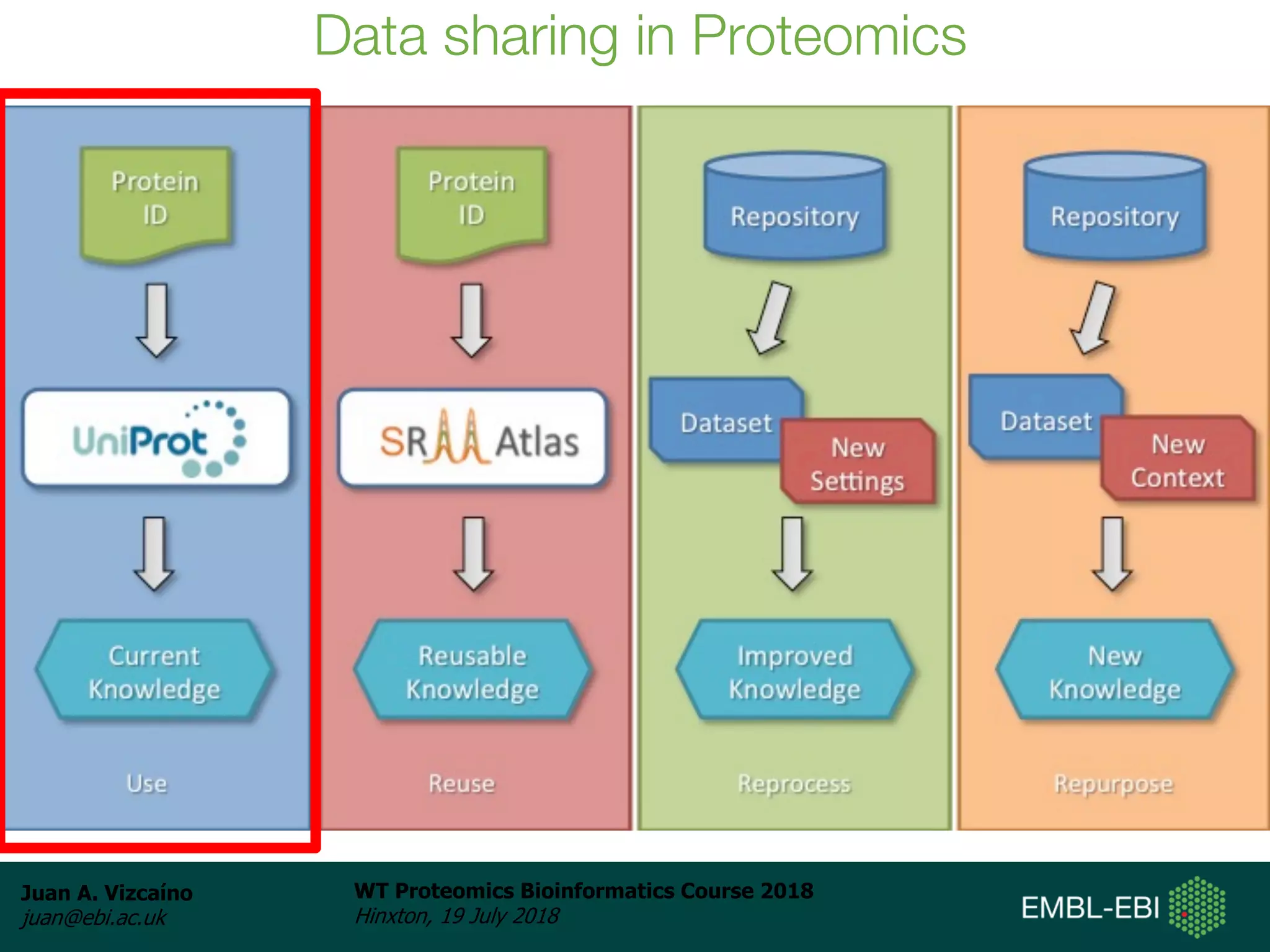
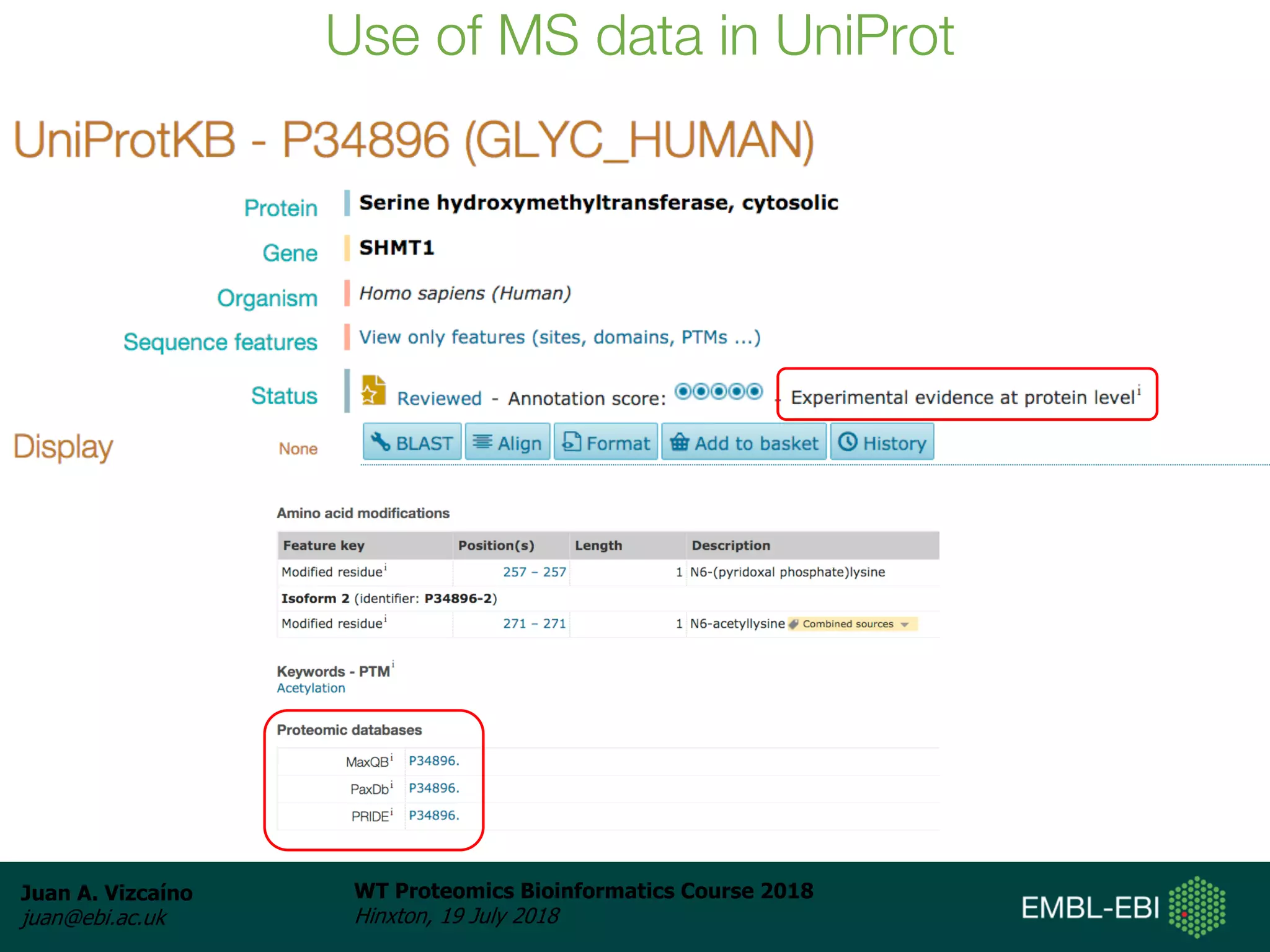
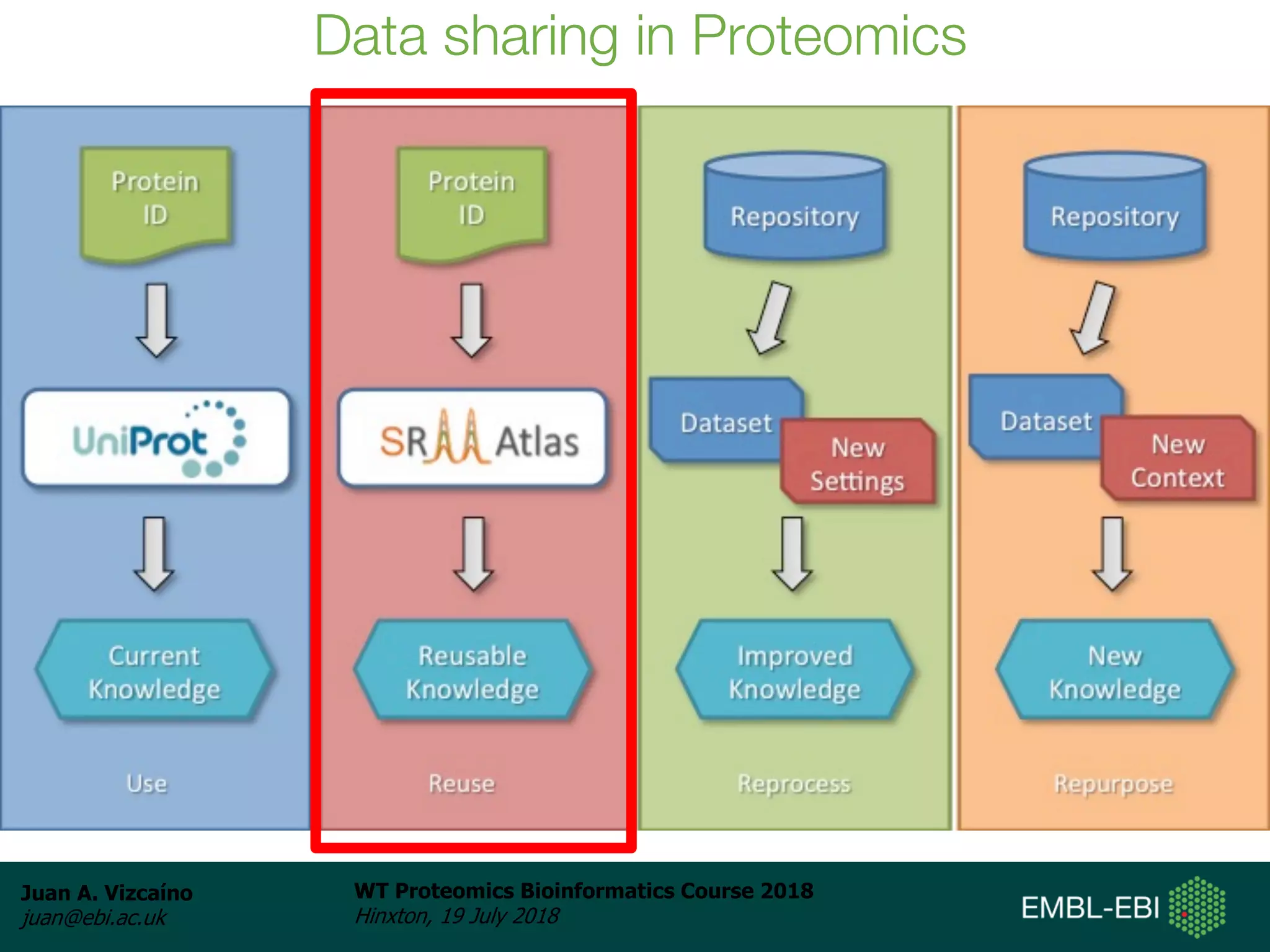
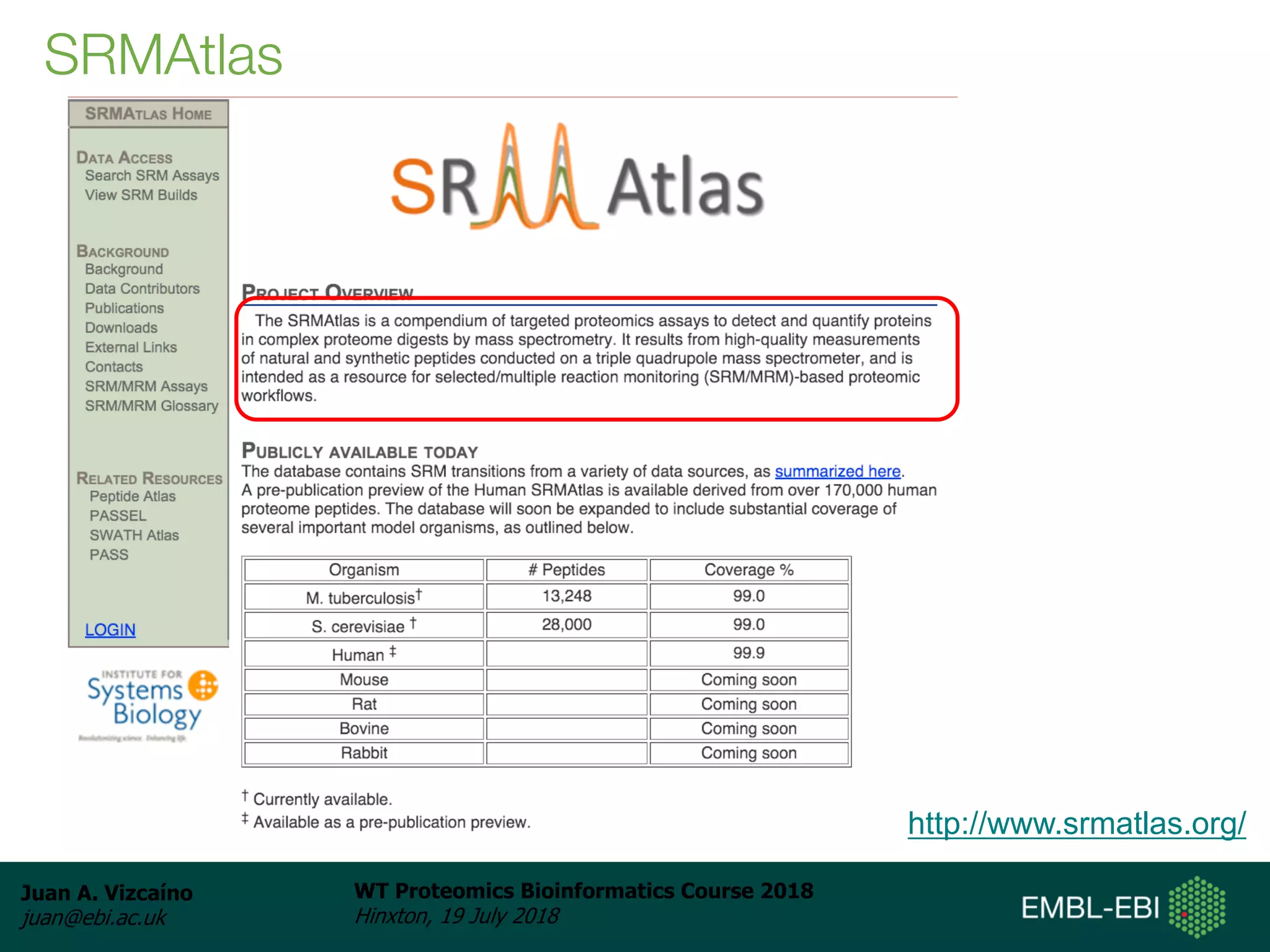
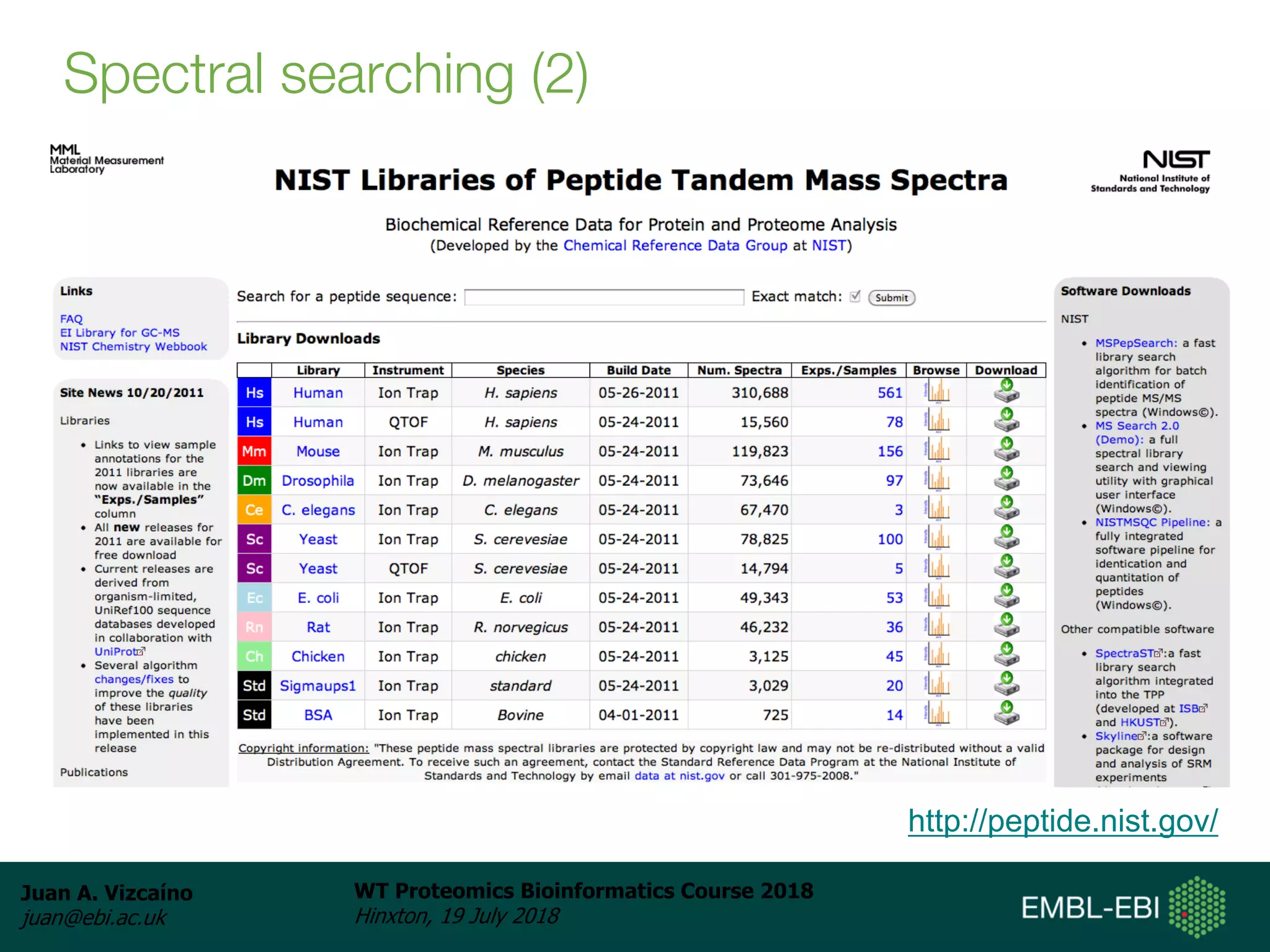
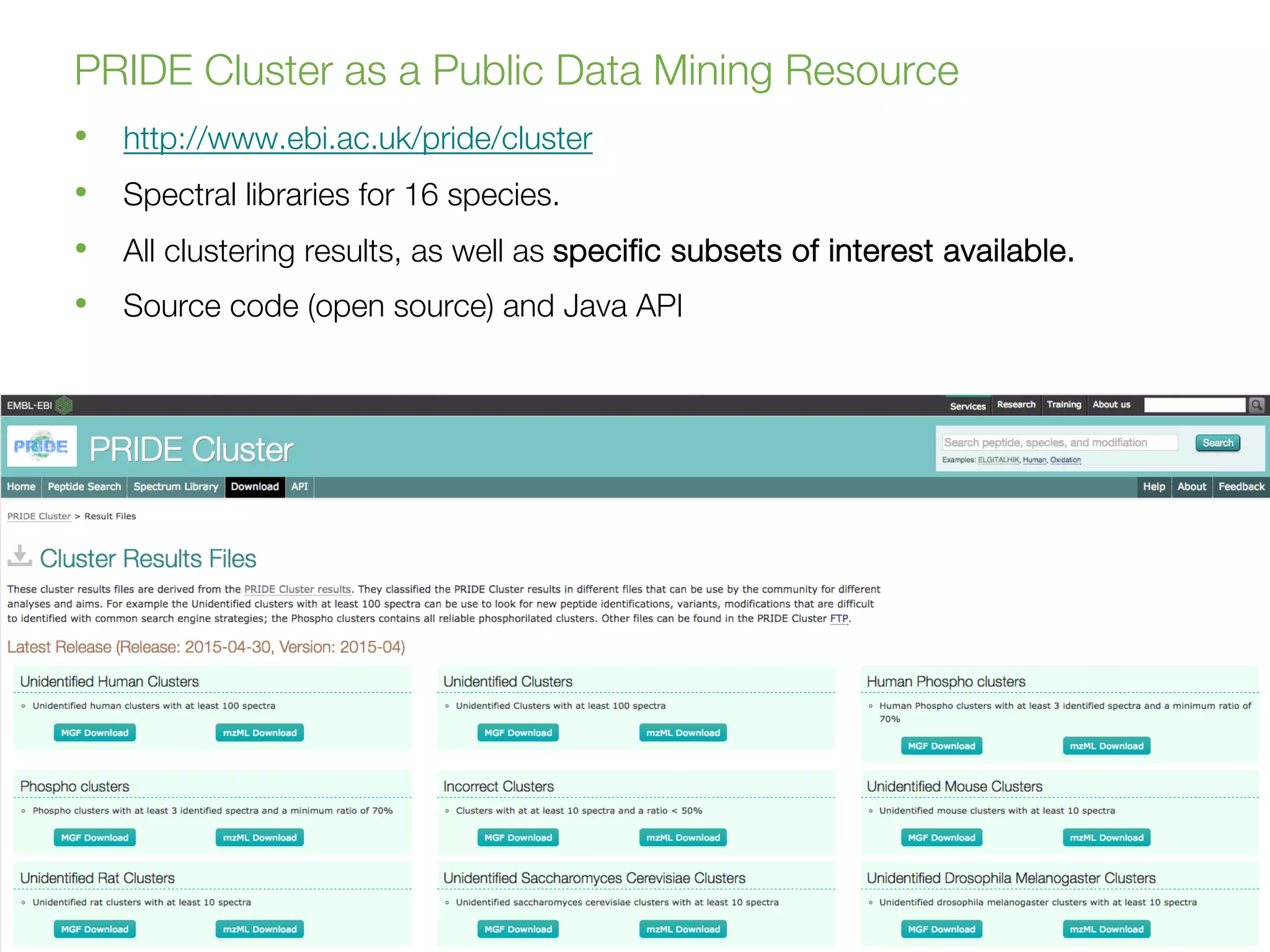
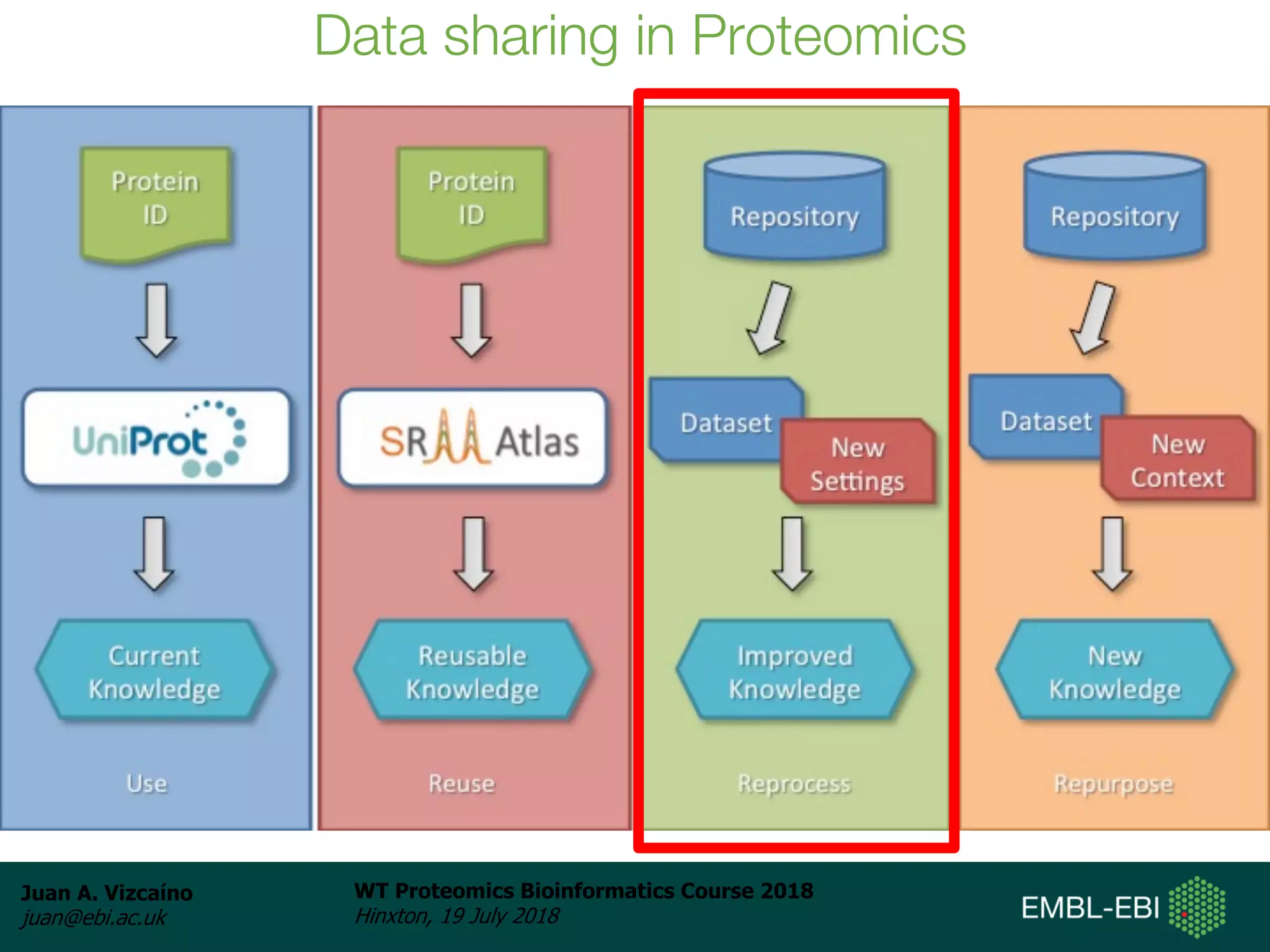
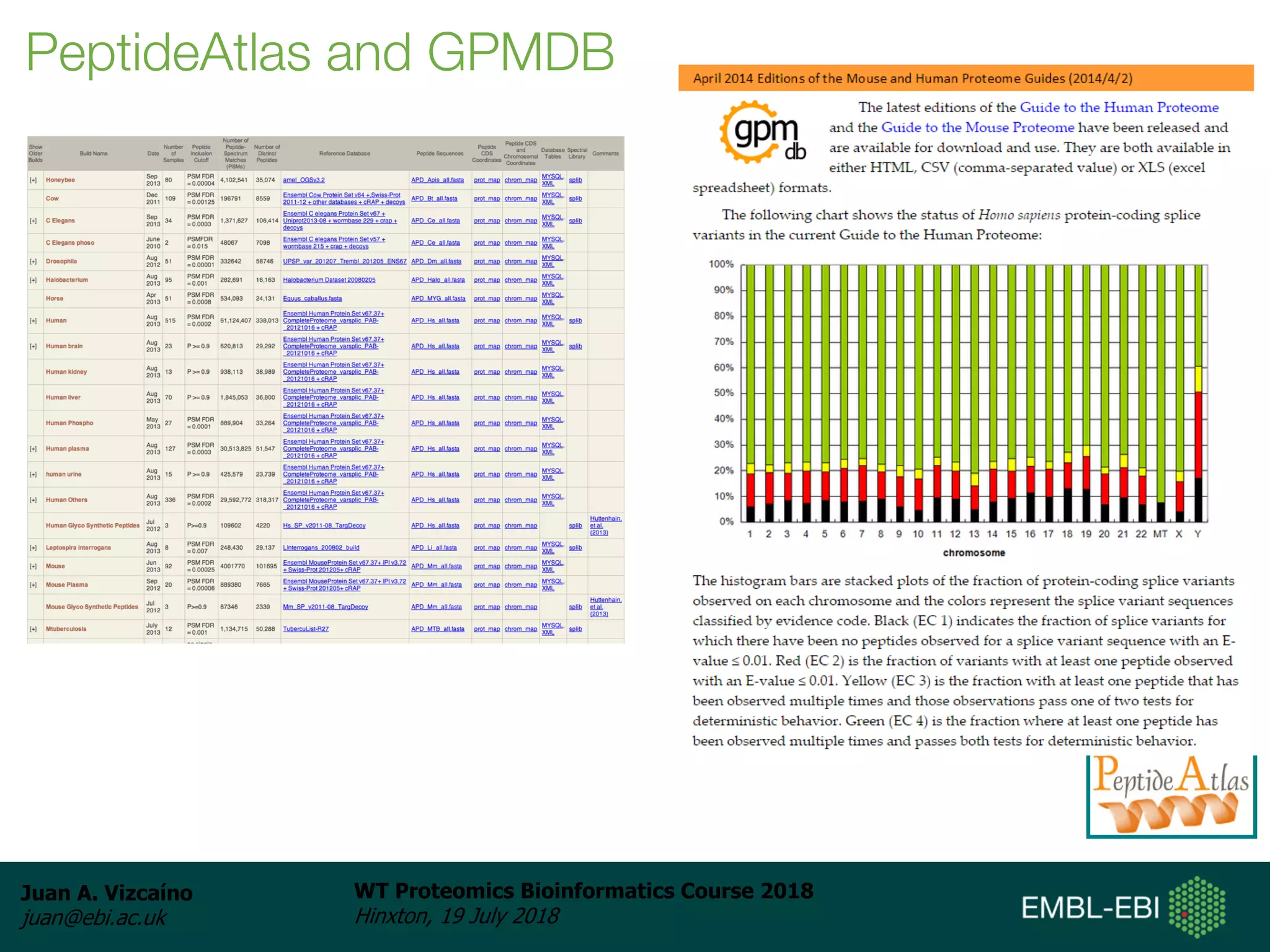
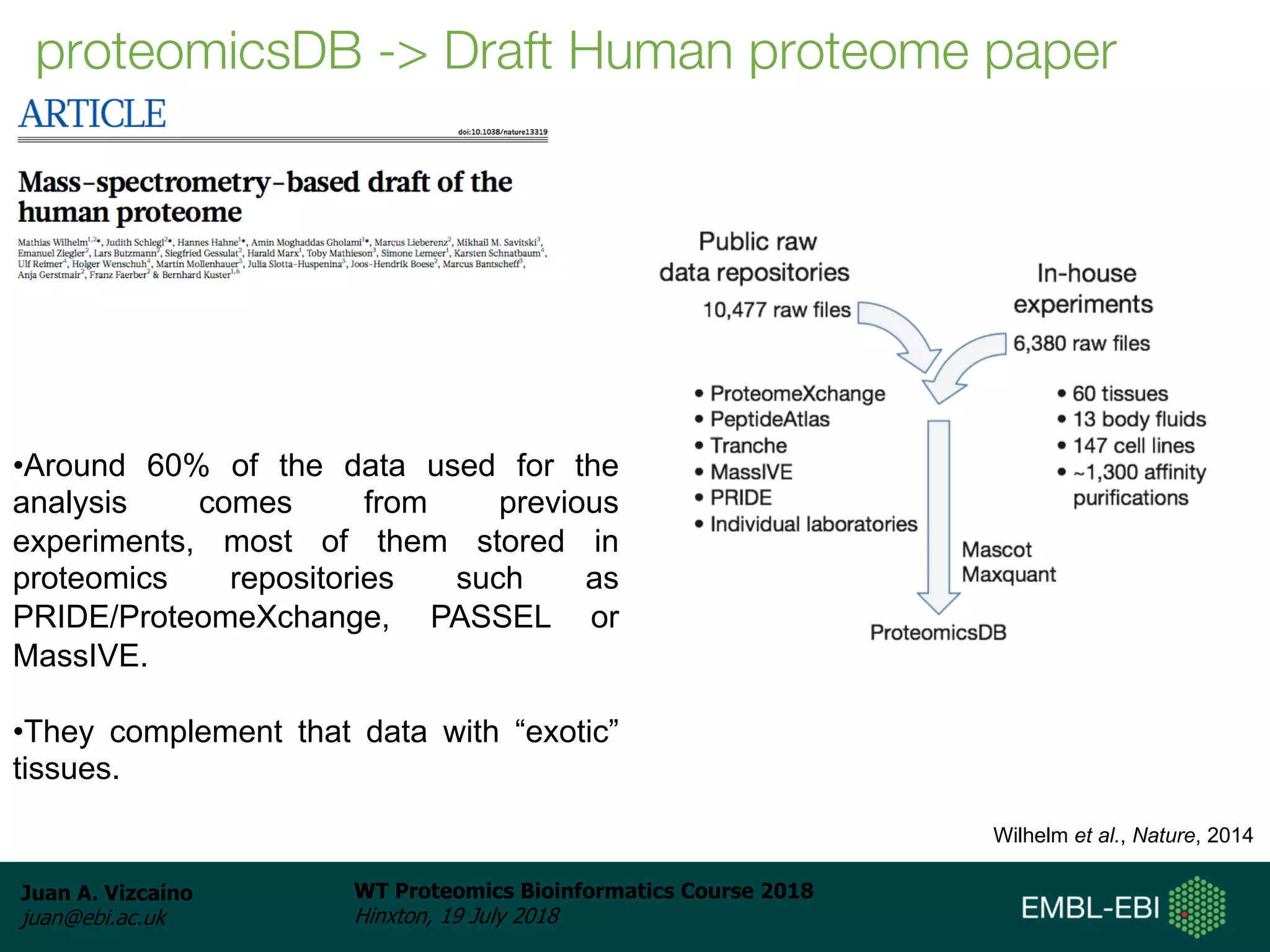
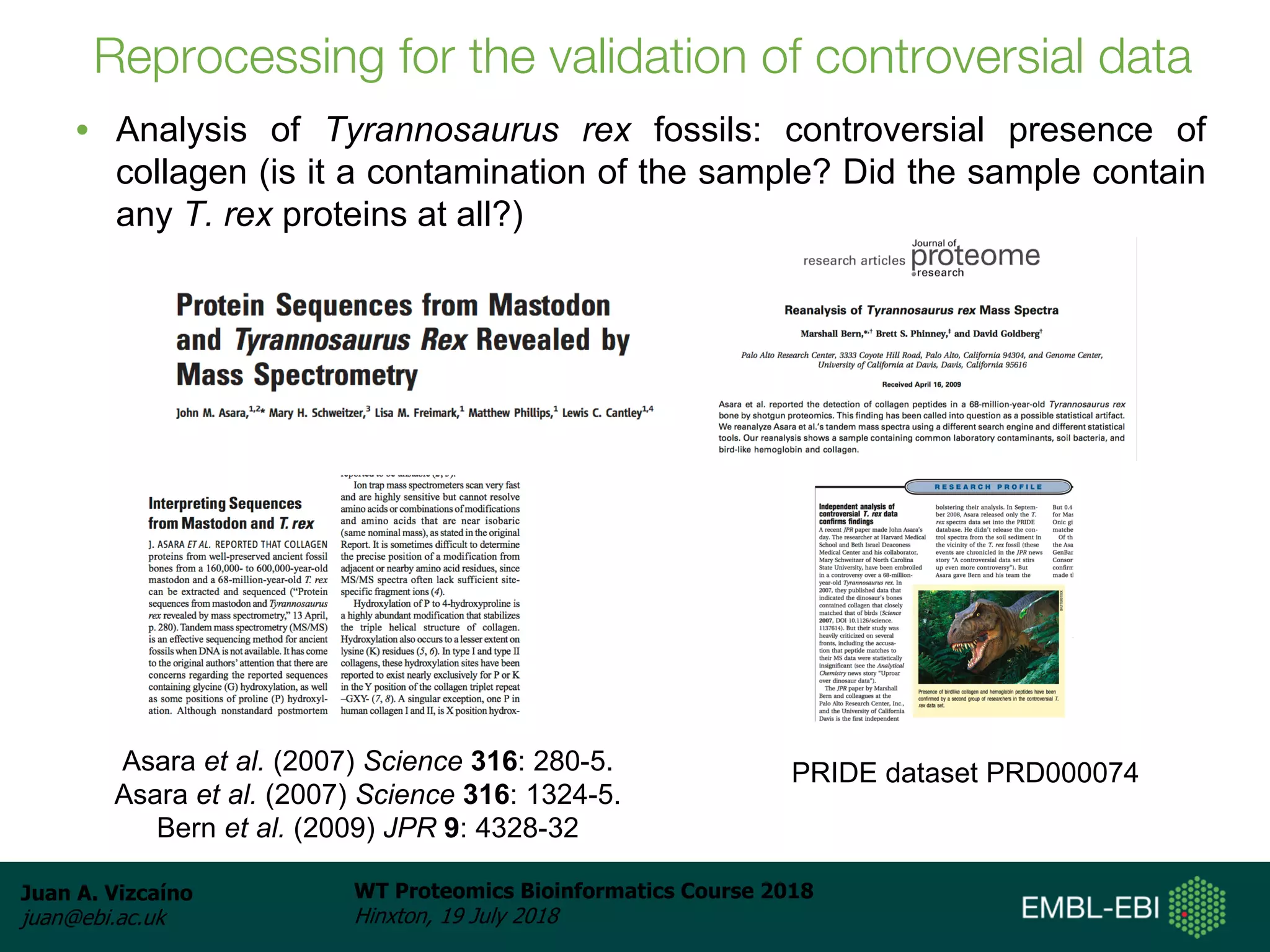
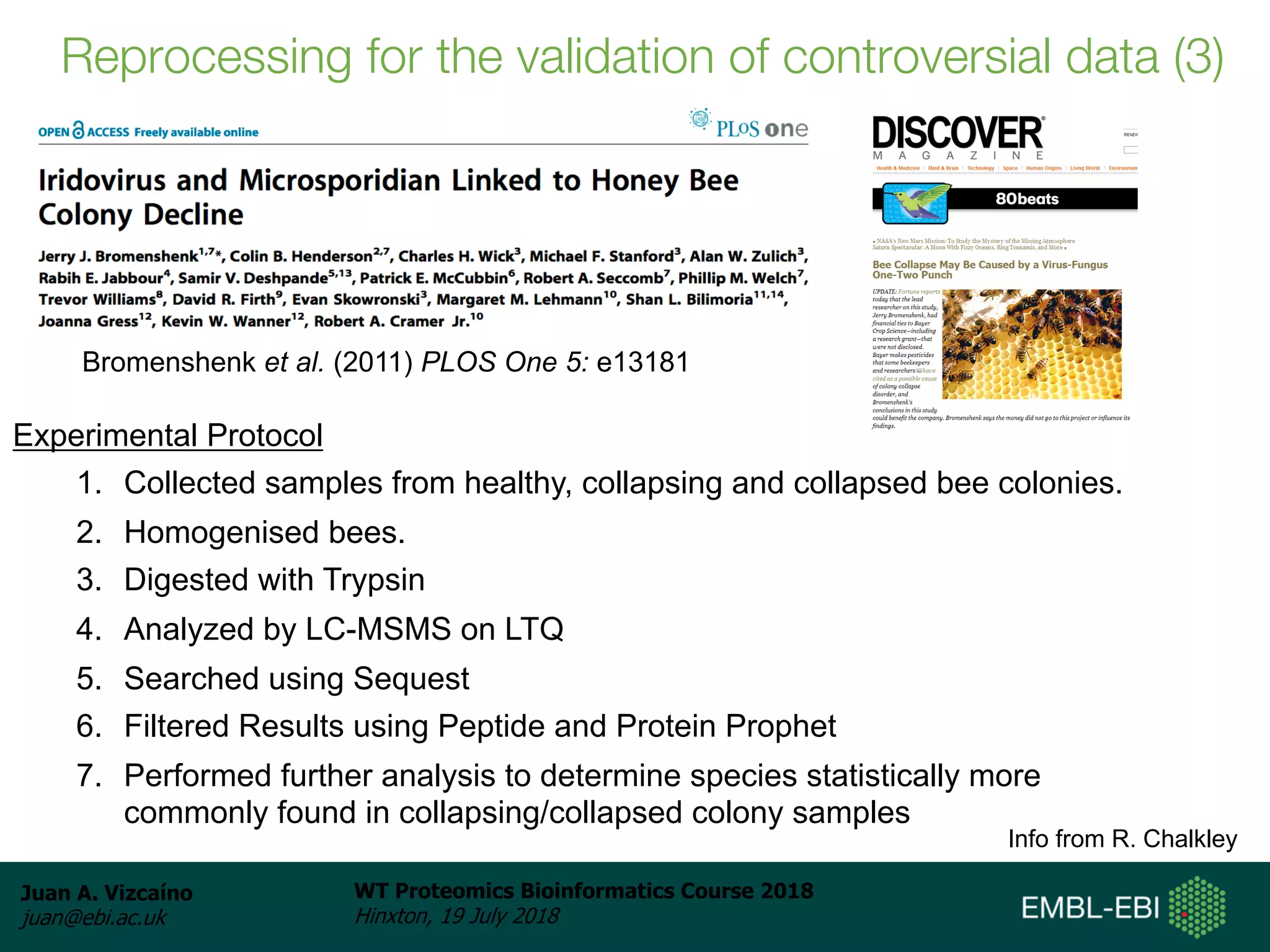
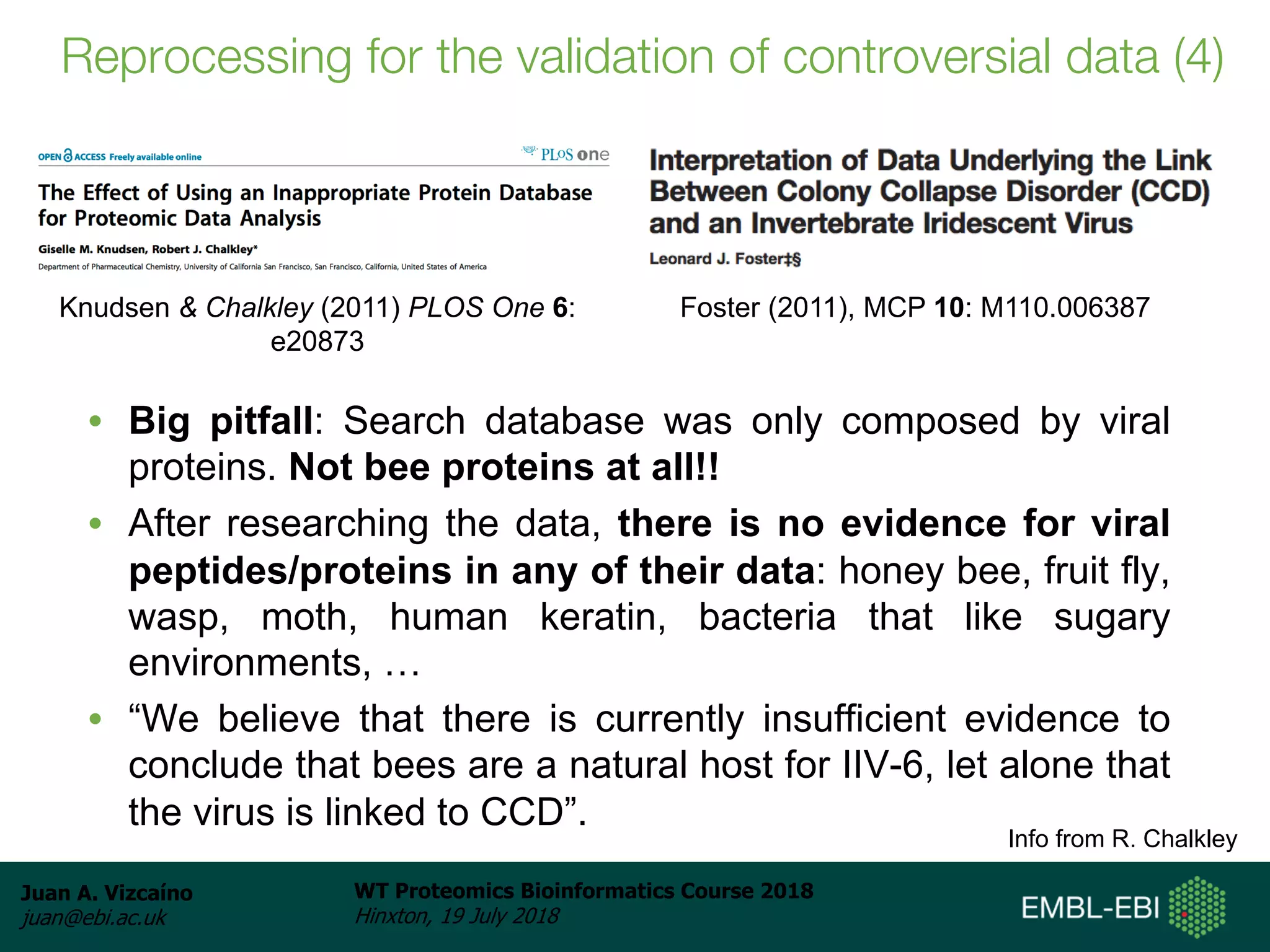
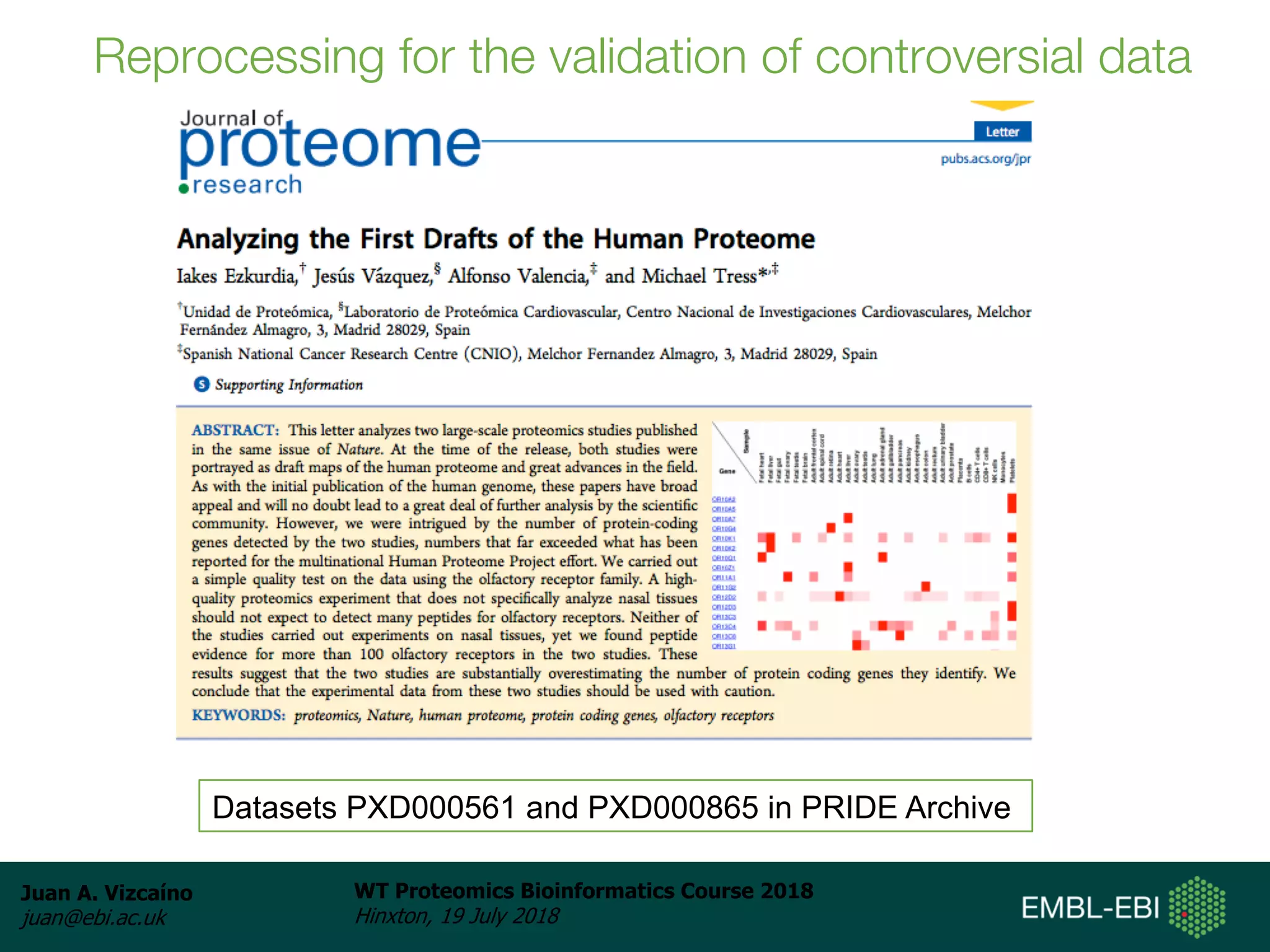
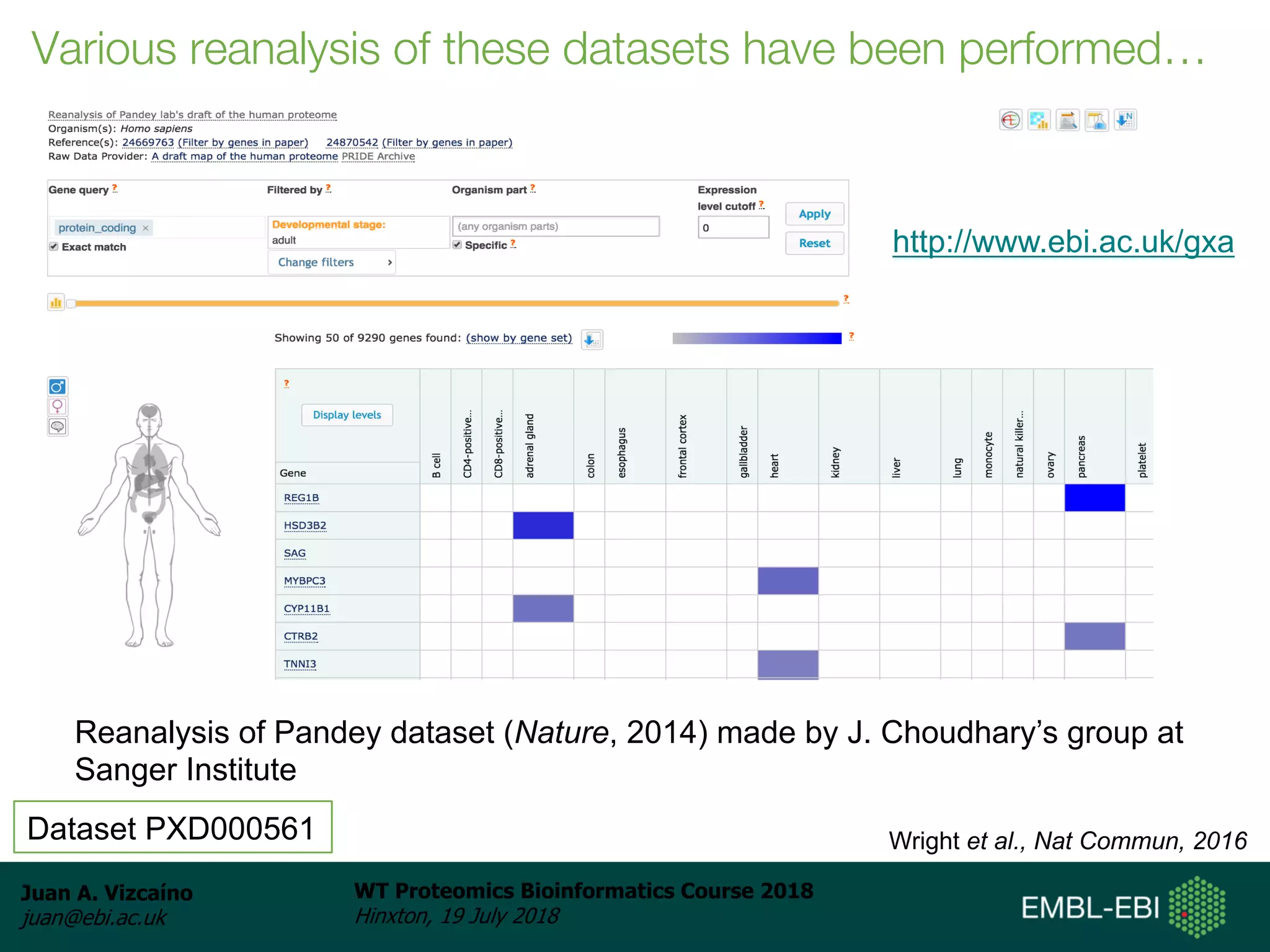
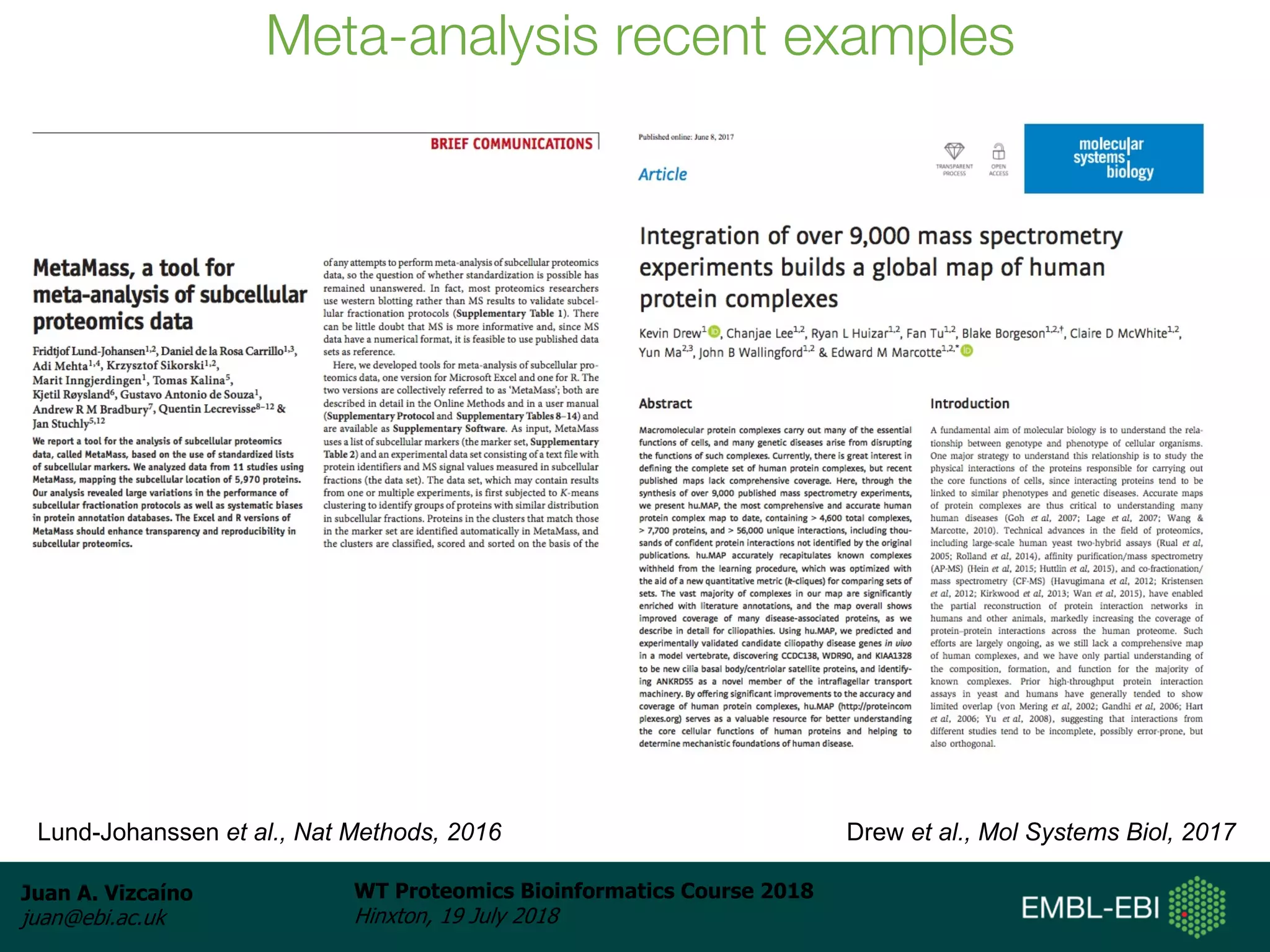
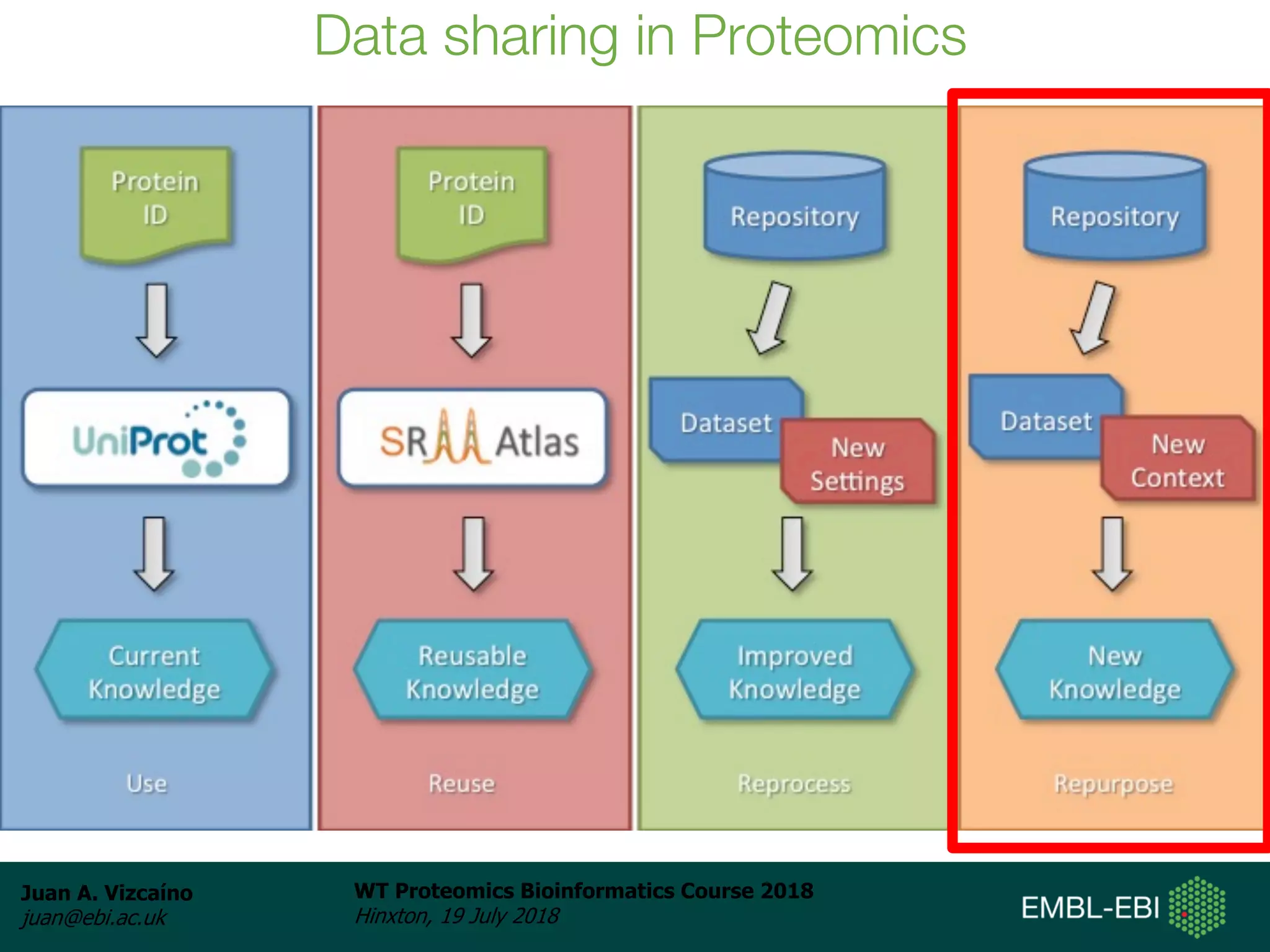
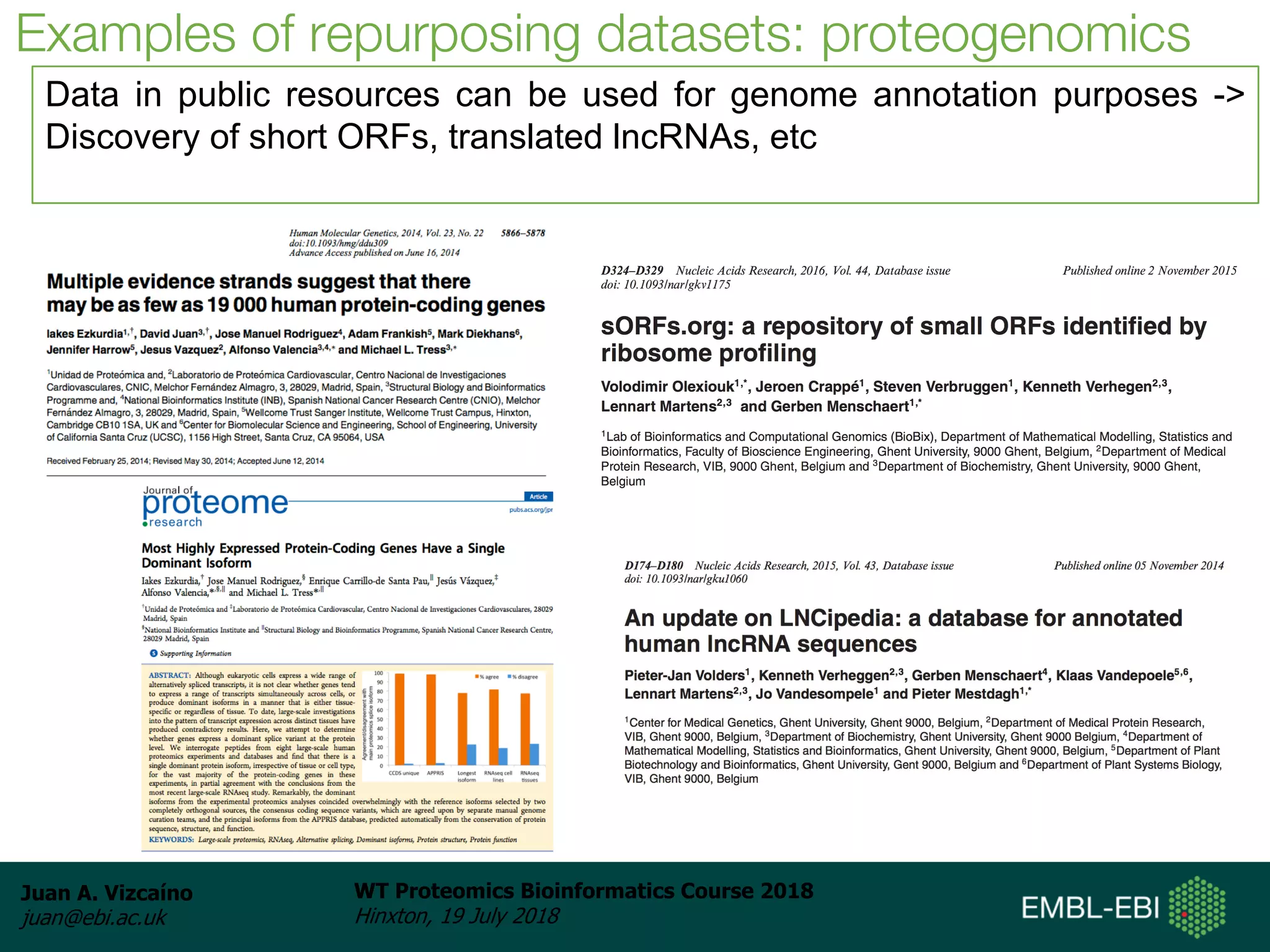
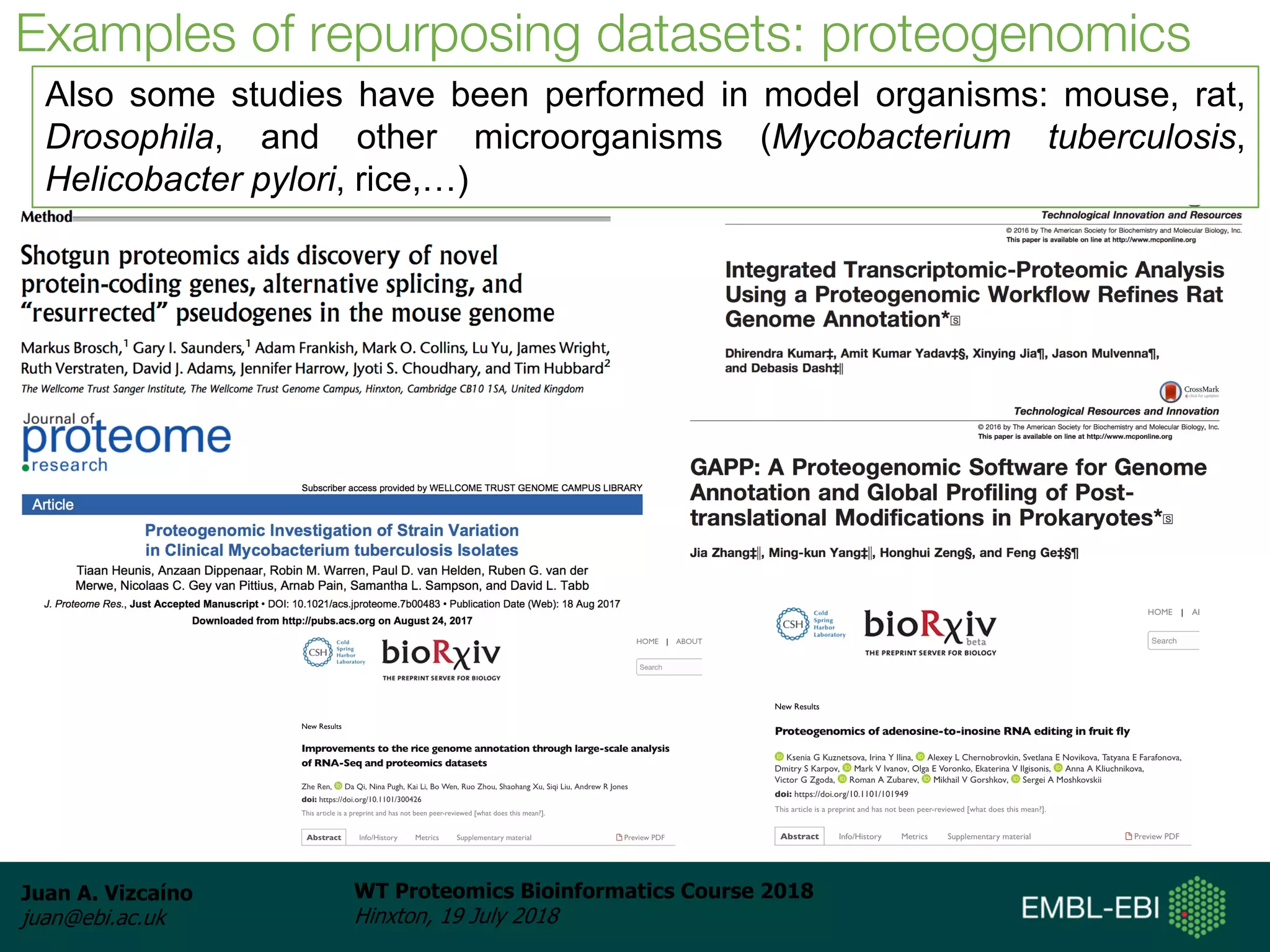
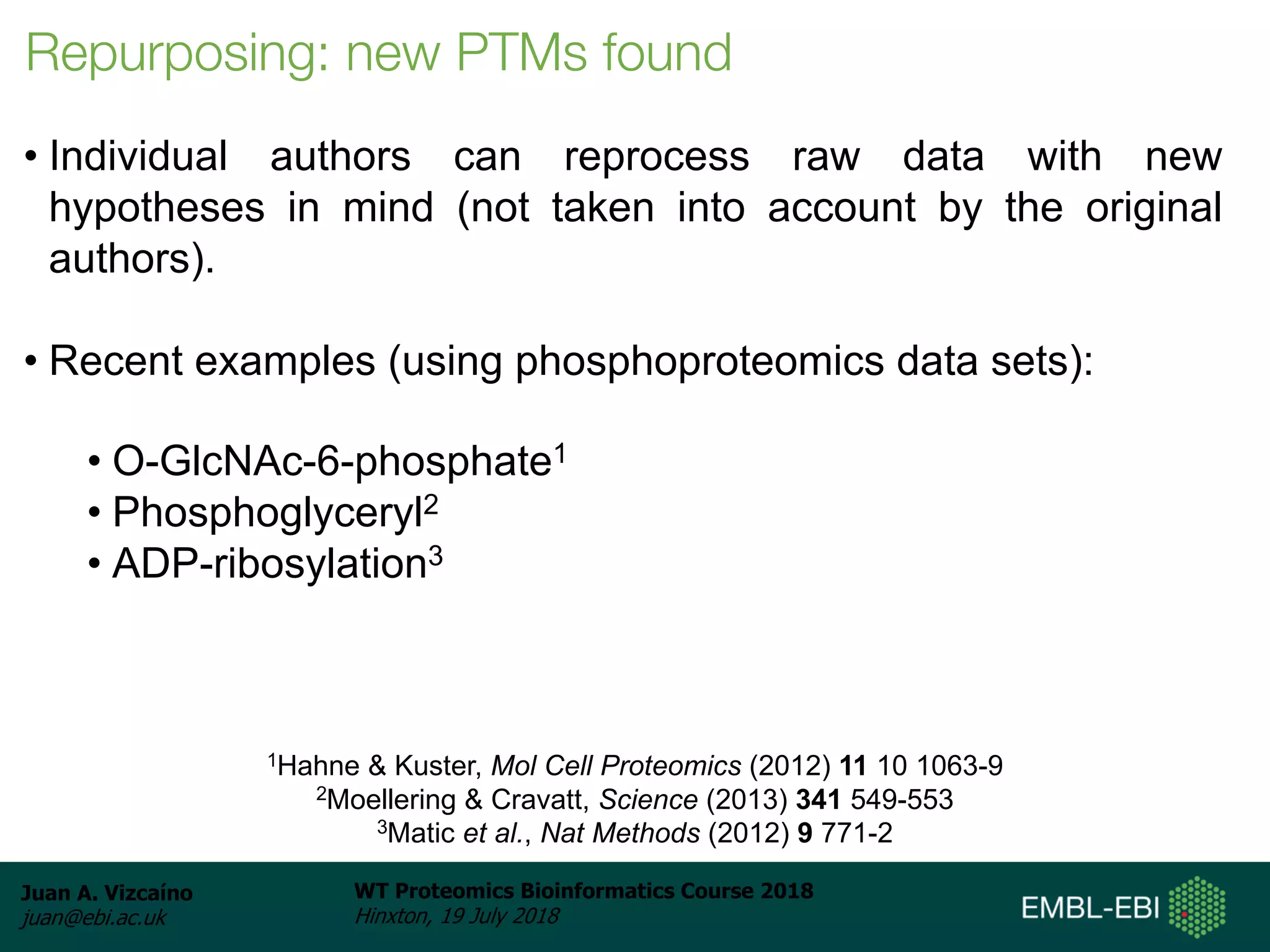
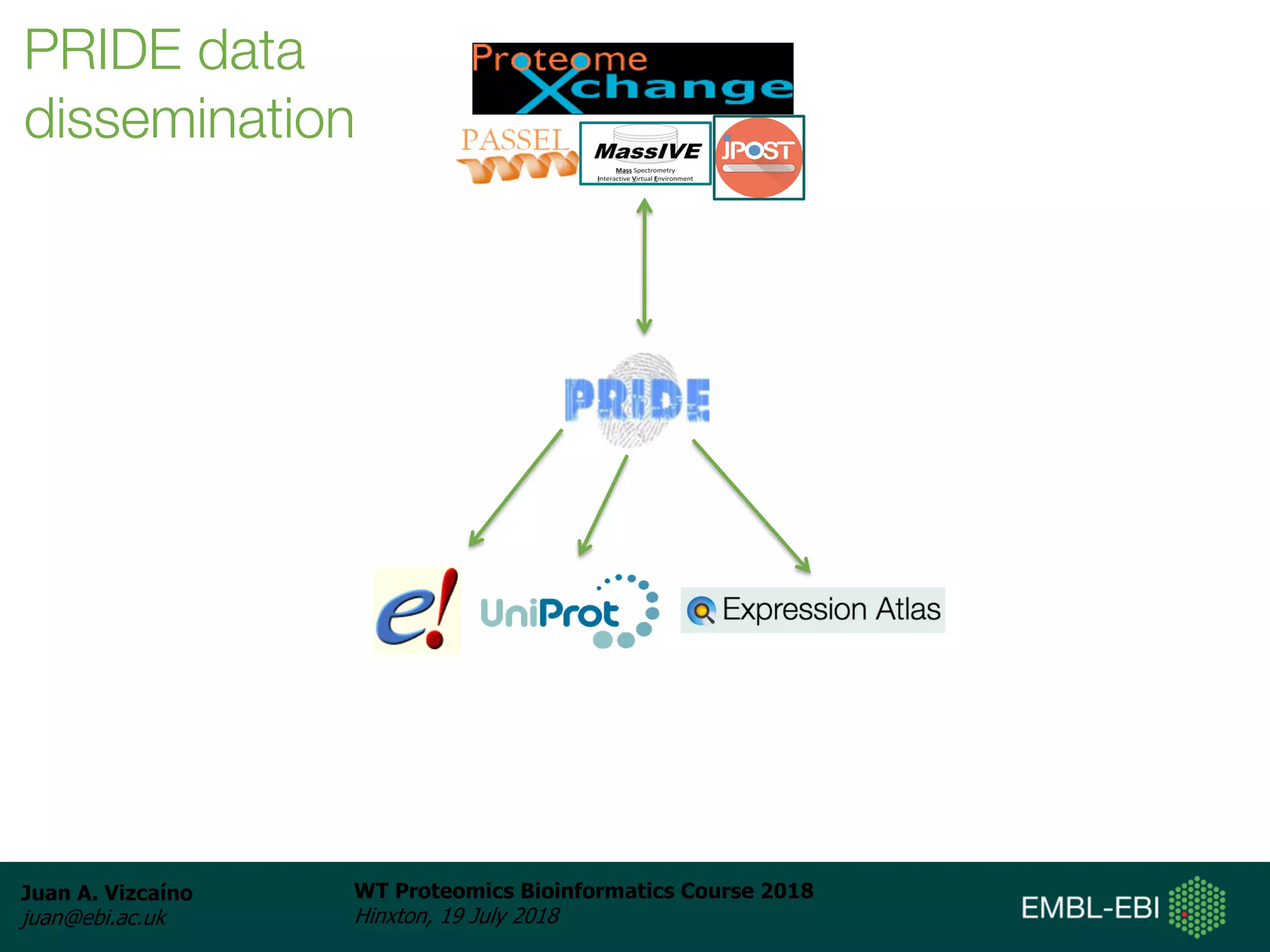
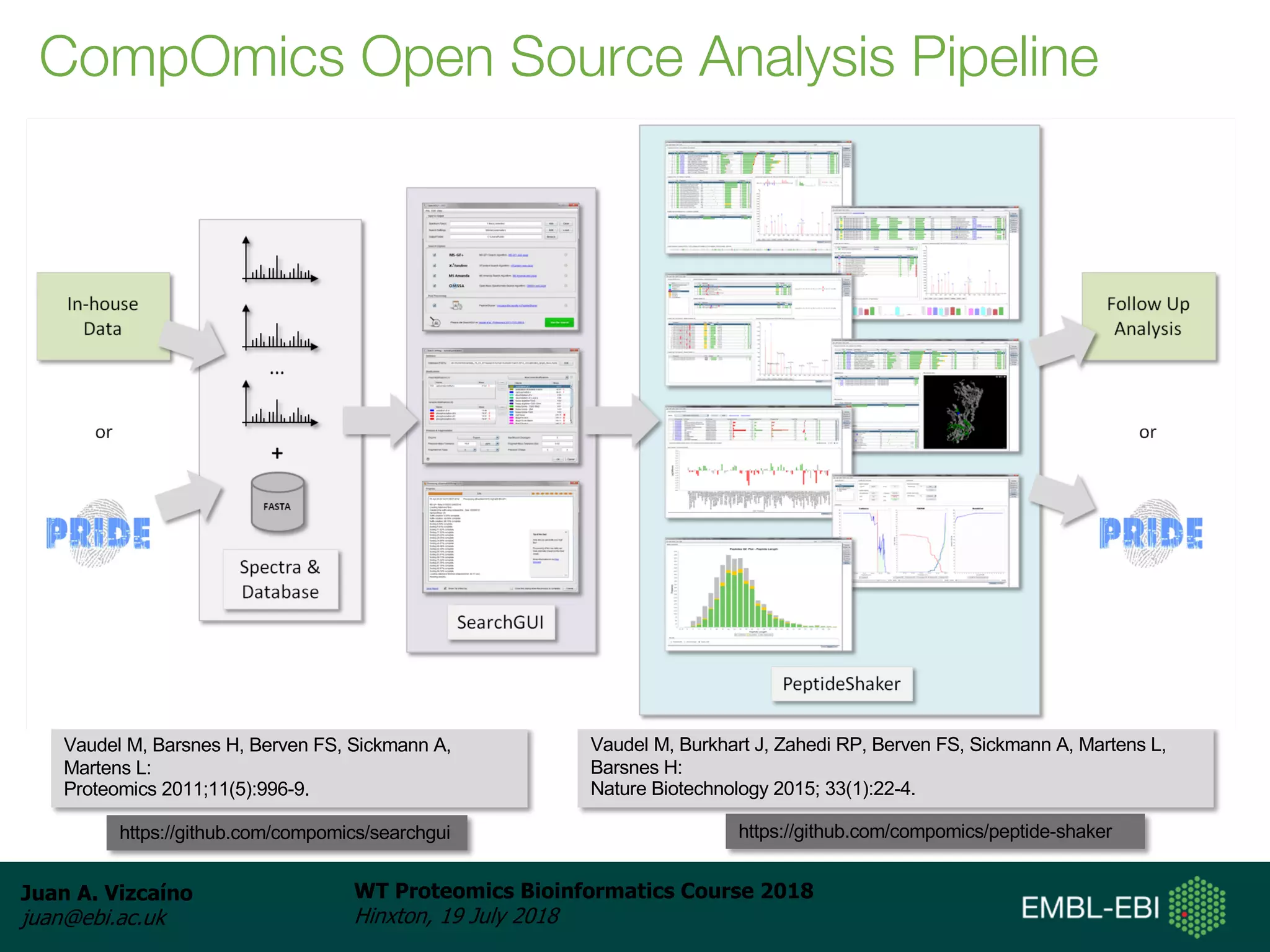
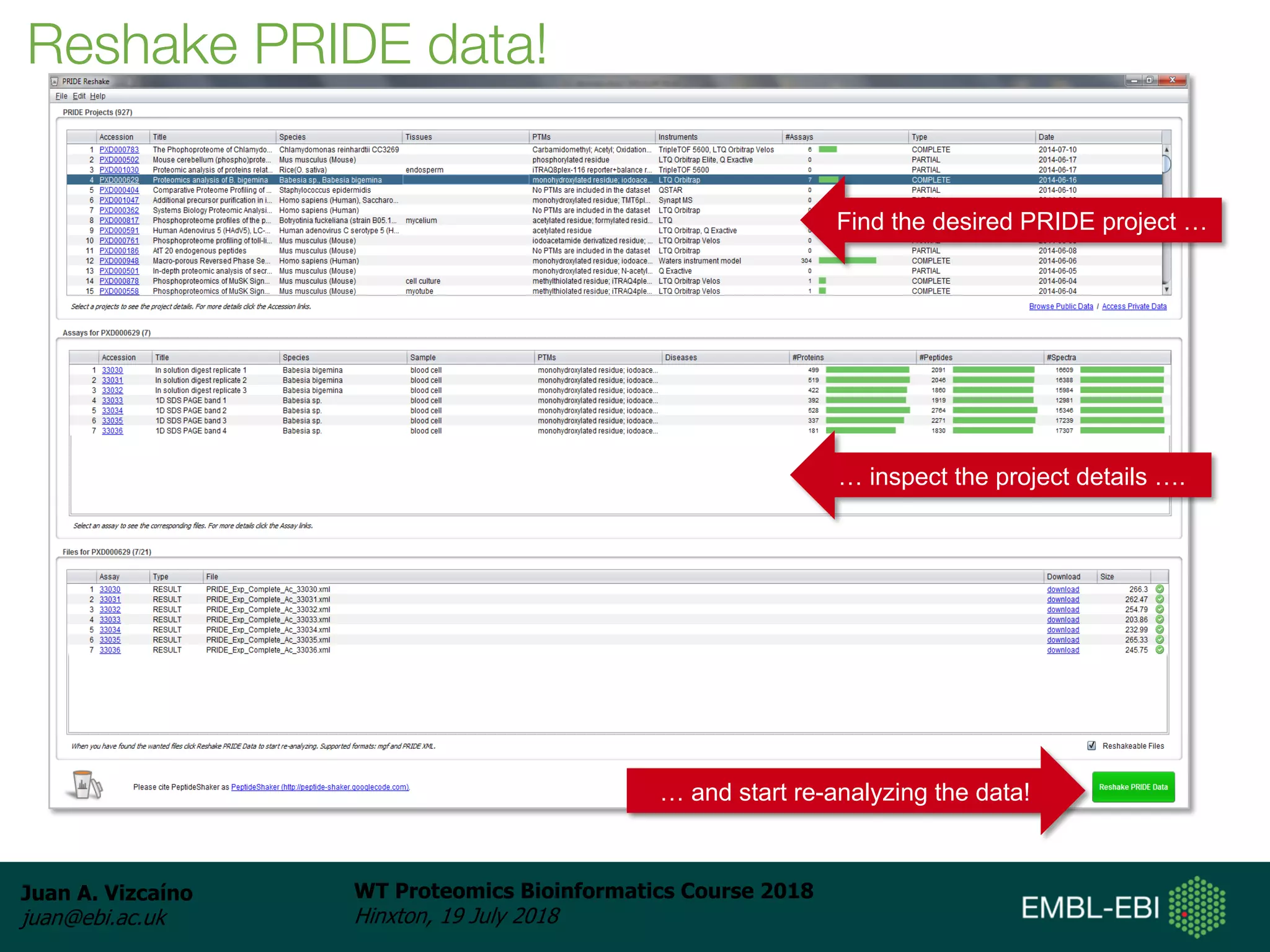
Dr. Juan Antonio Vizcaíno presented on the reuse of public proteomics data. The submission of proteomics datasets to repositories like PRIDE has increased dramatically in recent years. Downloads and reuse of data from PRIDE has also grown significantly, reaching 295 terabytes in 2017. Common ways researchers reuse public proteomics data include verifying published results, building spectral libraries, finding interesting datasets to reanalyze for new discoveries, and benchmarking new algorithms. Data sharing allows information to be extracted and reused in new experiments, advancing protein knowledge in areas like UniProt and neXtProt databases.