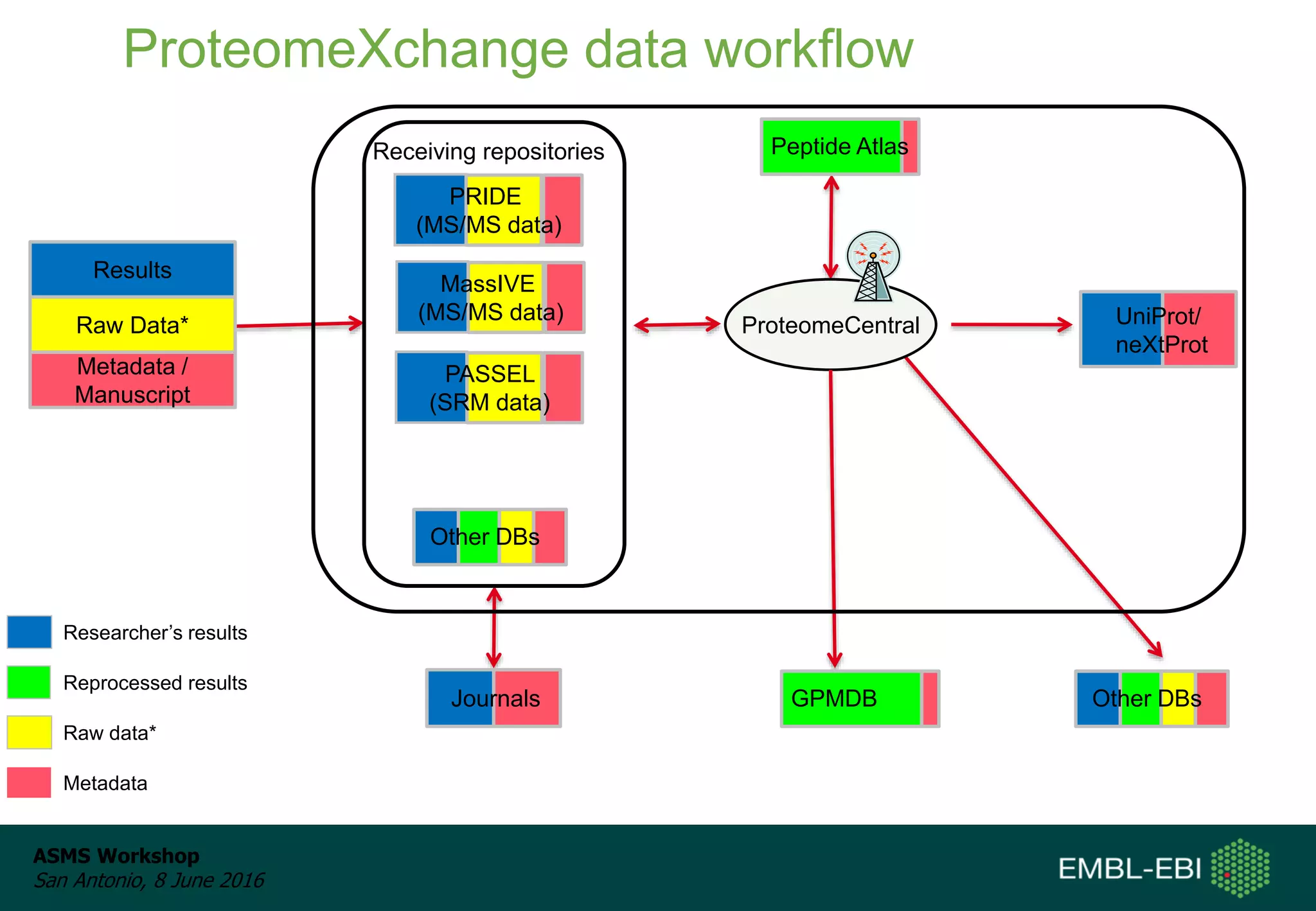
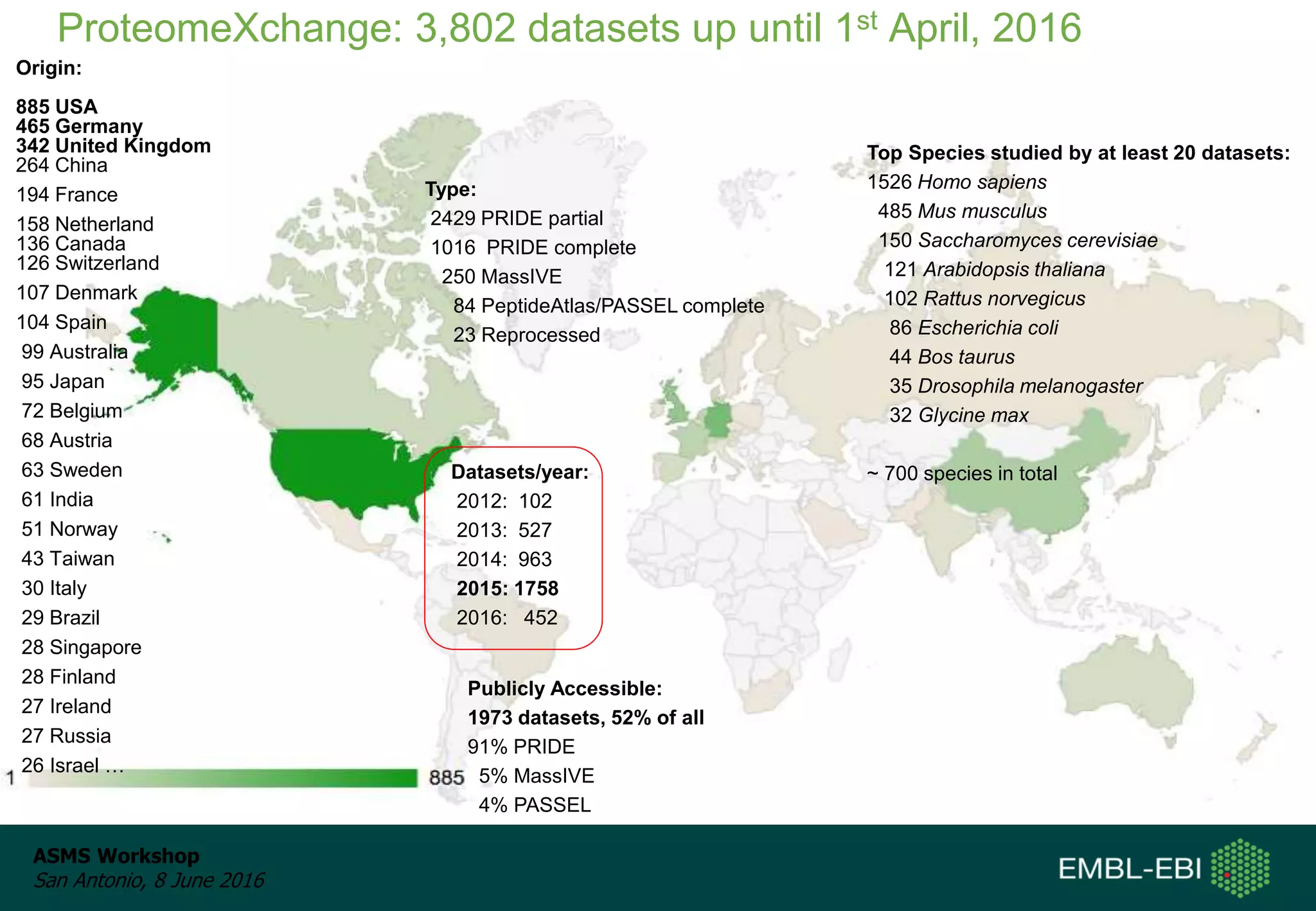
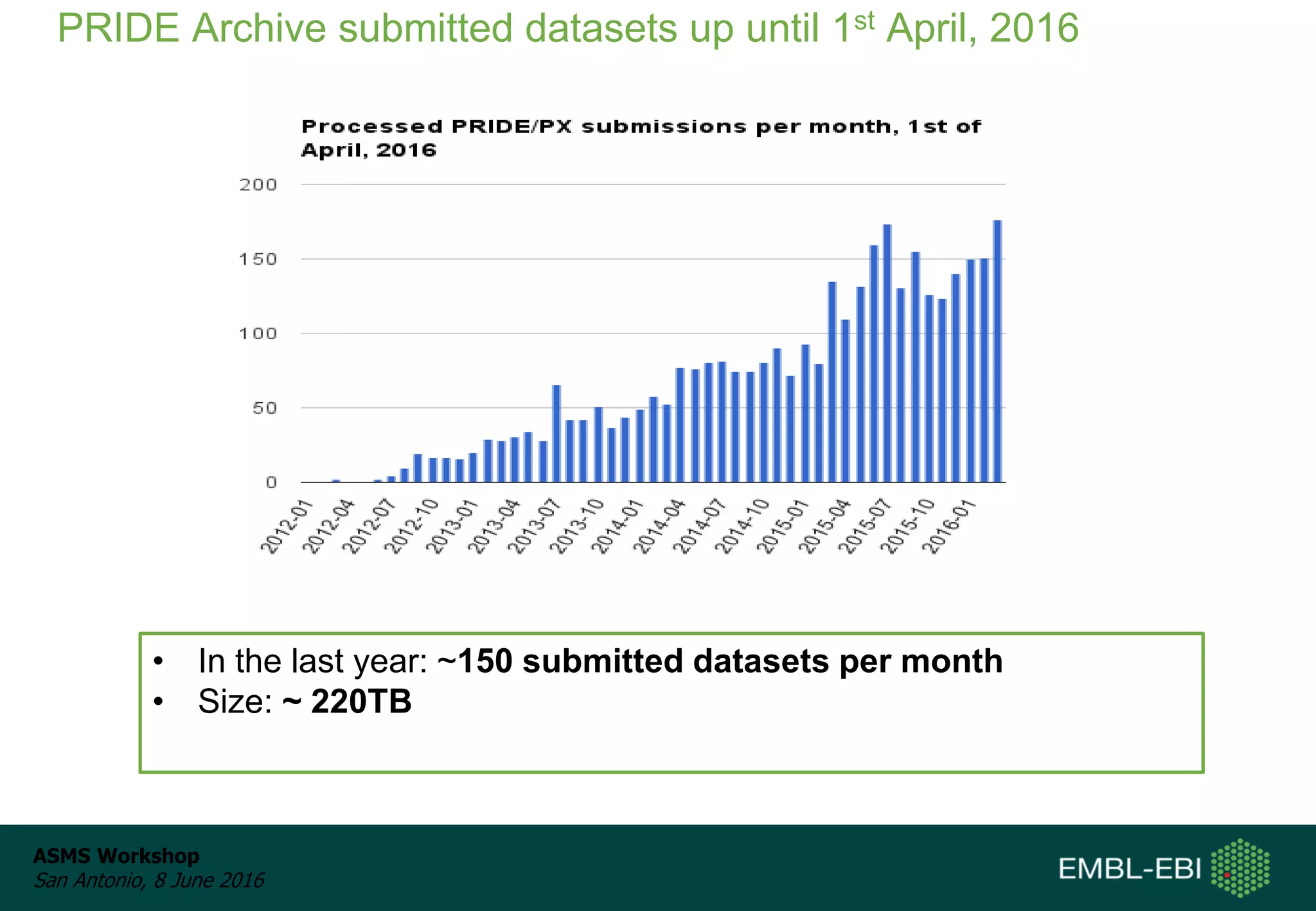
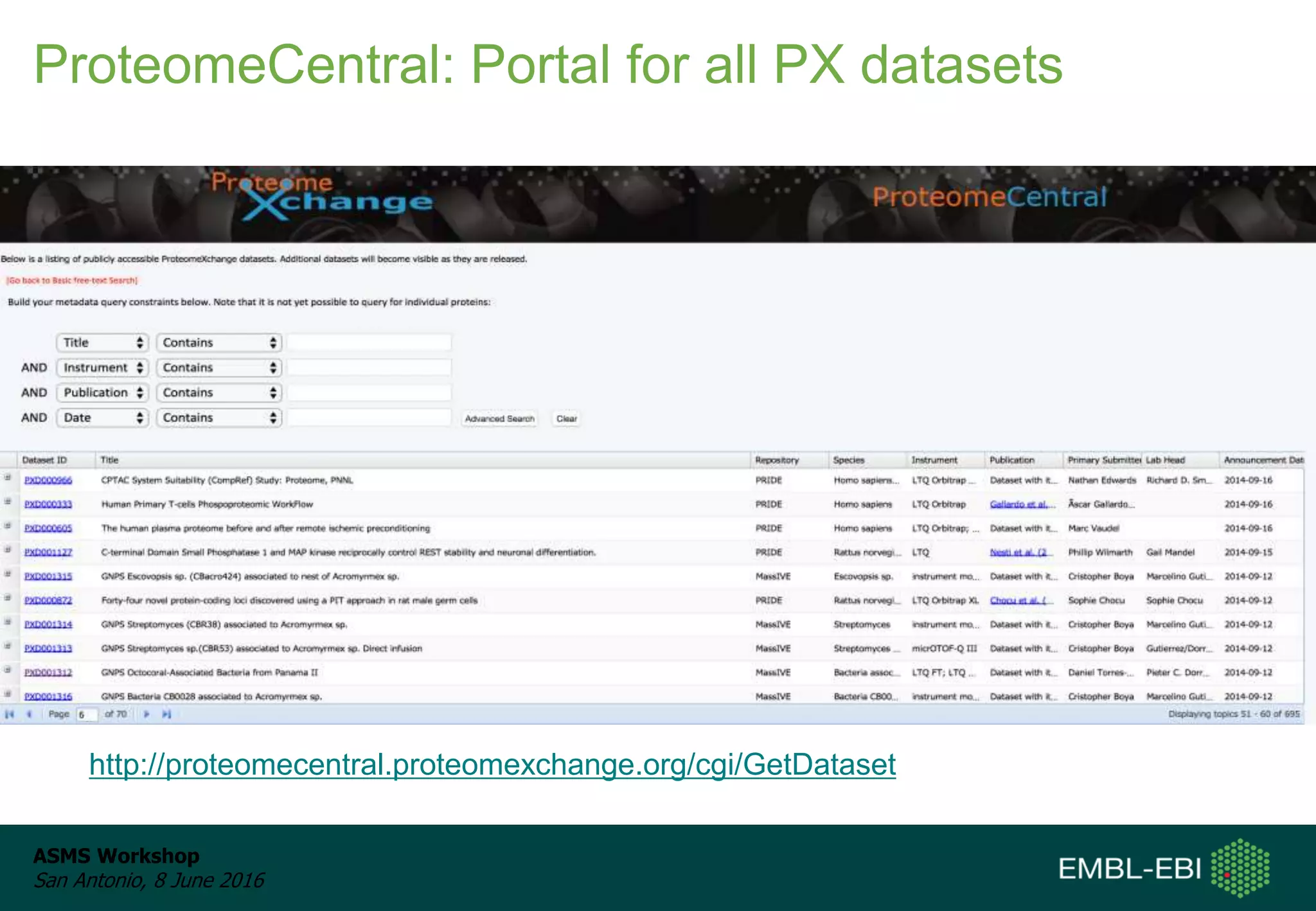
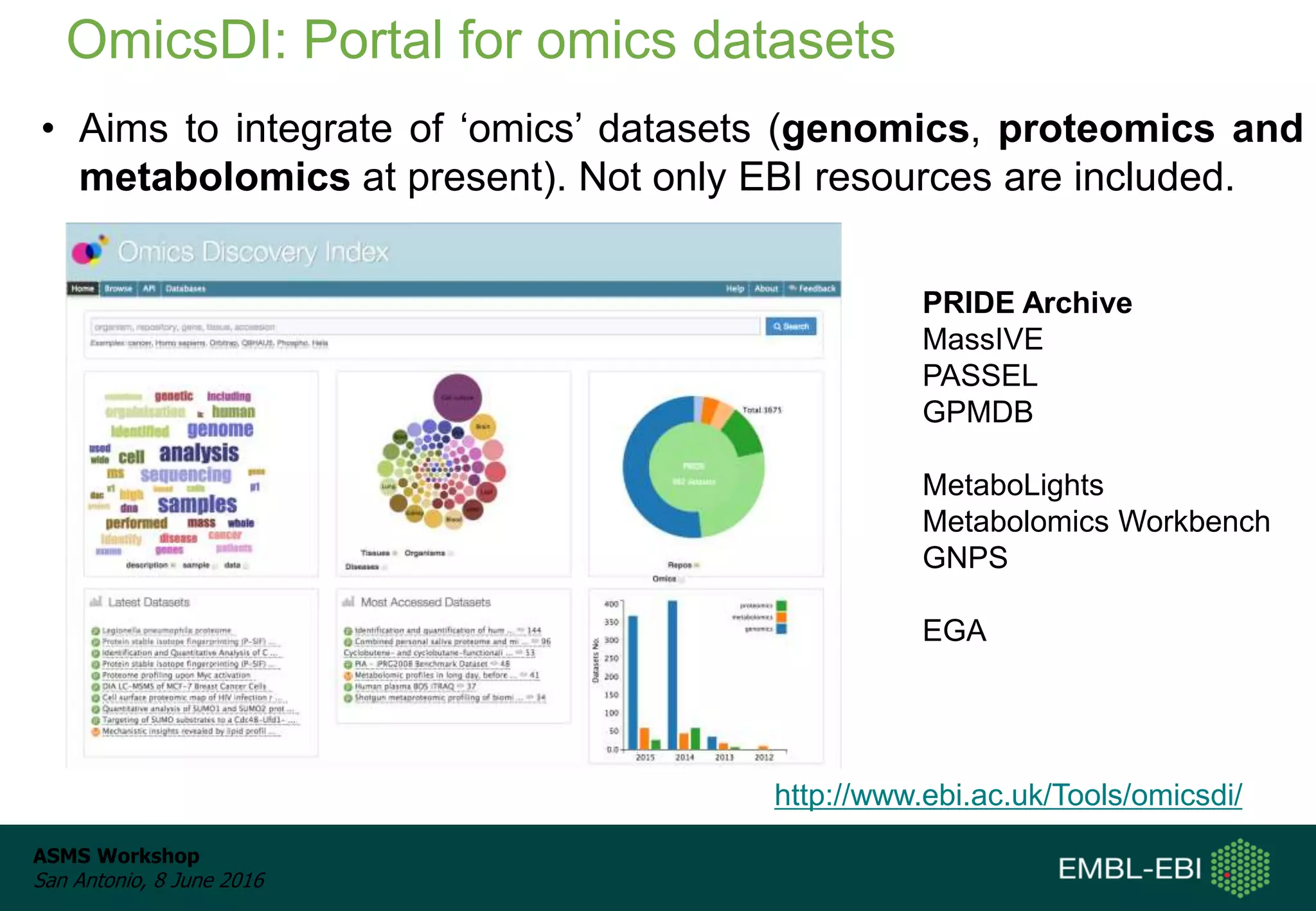
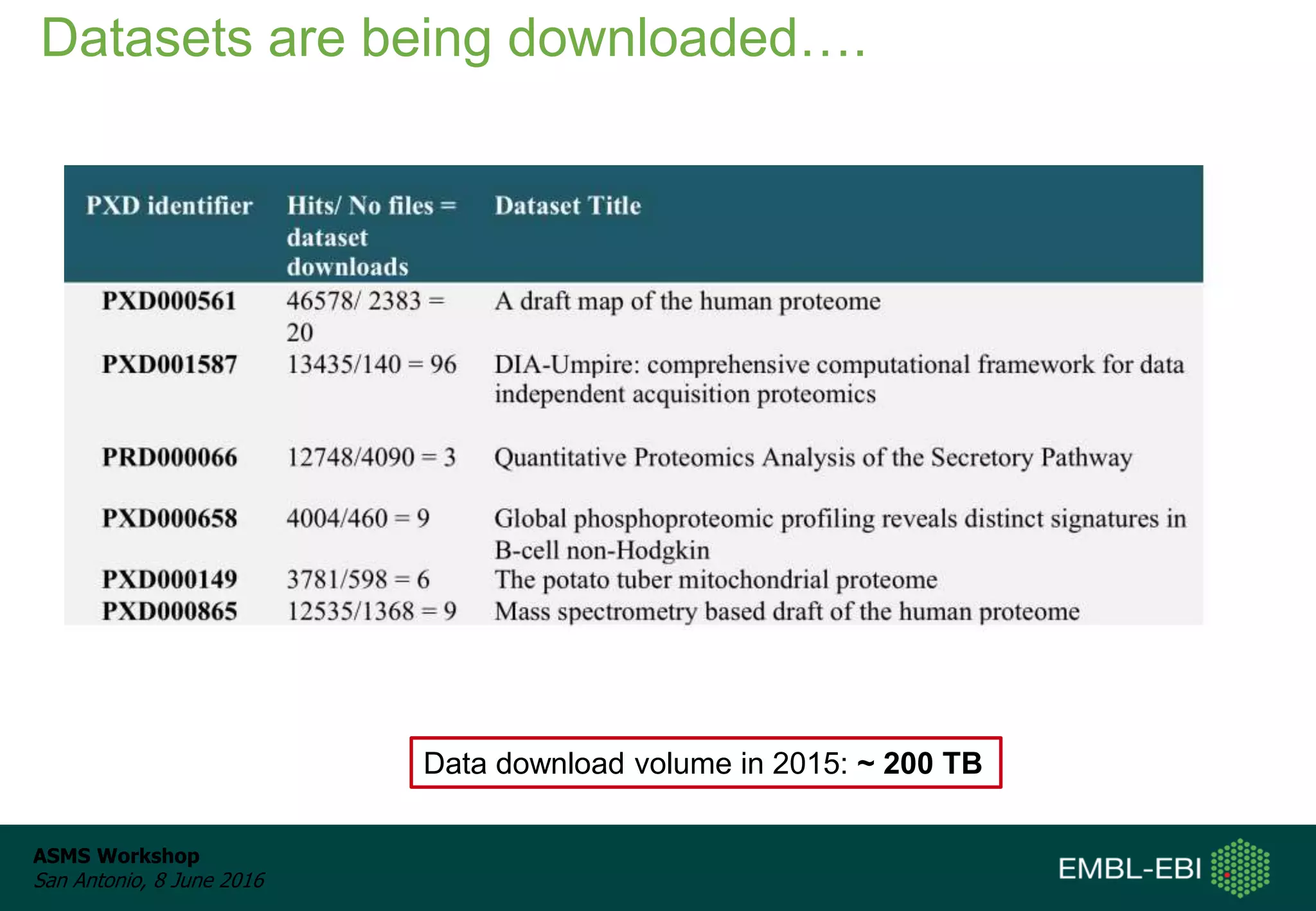
The ProteomeXchange Consortium aims to allow standard data submission and dissemination between major proteomics repositories, including PeptideAtlas, PRIDE, and MassIVE. It establishes a common identifier space (PXD IDs) and supports workflows for MS/MS and SRM data submitted from any experimental approach. Since 2012, over 3,800 datasets have been submitted from over 700 species, with over 1,900 publicly accessible. Submissions have grown significantly each year, and data downloads for reuse are also increasing. The goal is to make data sharing easier for researchers.