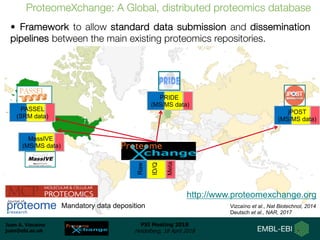
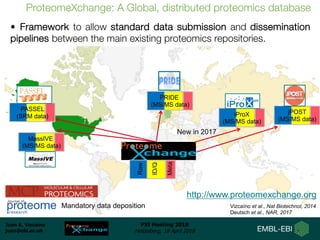
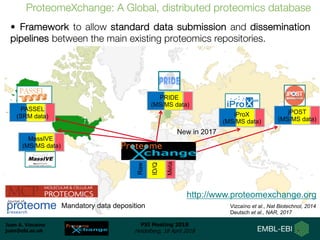
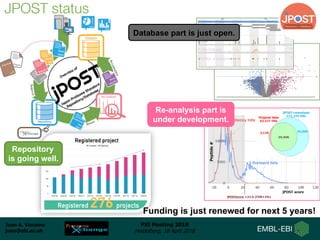
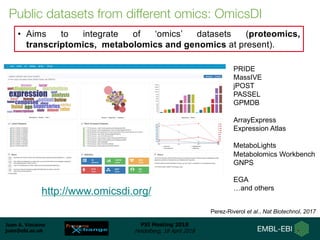
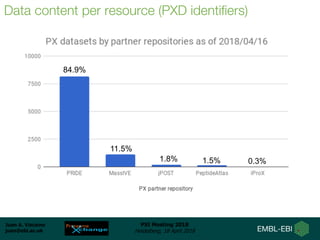
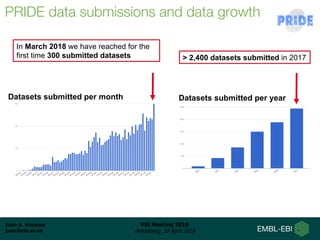
1) ProteomeXchange is a global database containing proteomics data from several repositories including PRIDE, MassIVE, and jPOST.
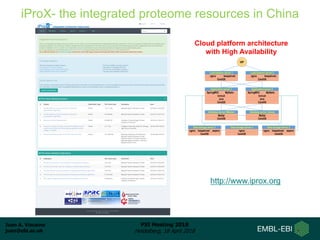
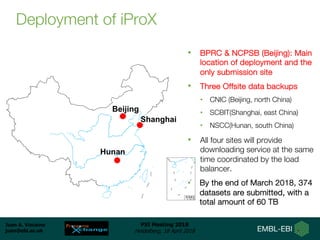
2) A new member, iProX, joined in 2017 and contains over 60 terabytes of data from China.
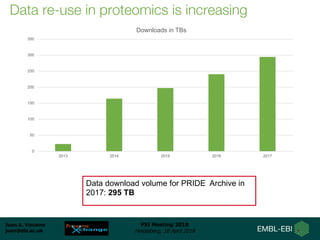
3) Usage of ProteomeXchange data is increasing, with PRIDE downloads growing from 50 terabytes in 2013 to over 295 terabytes in 2017.