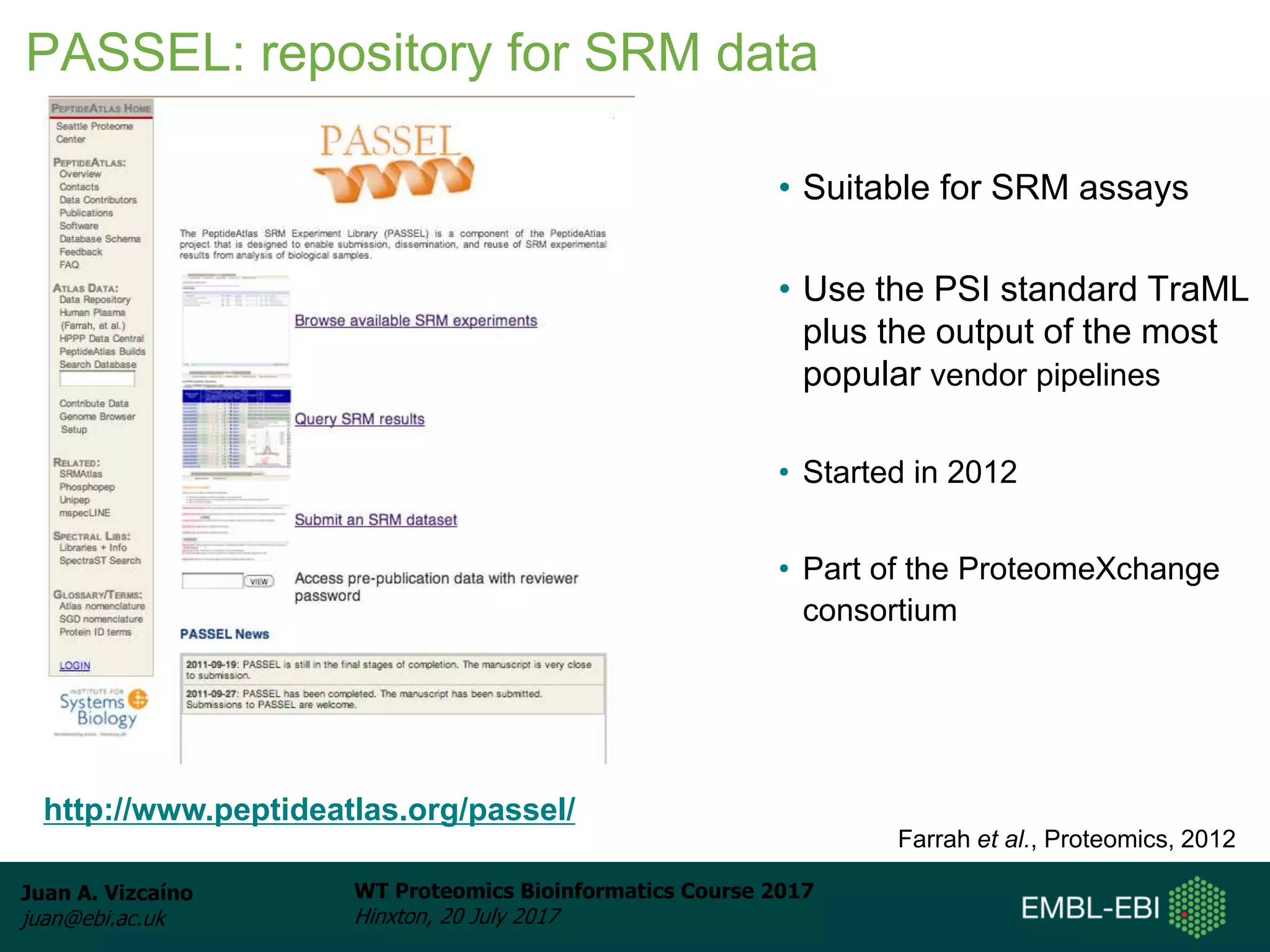
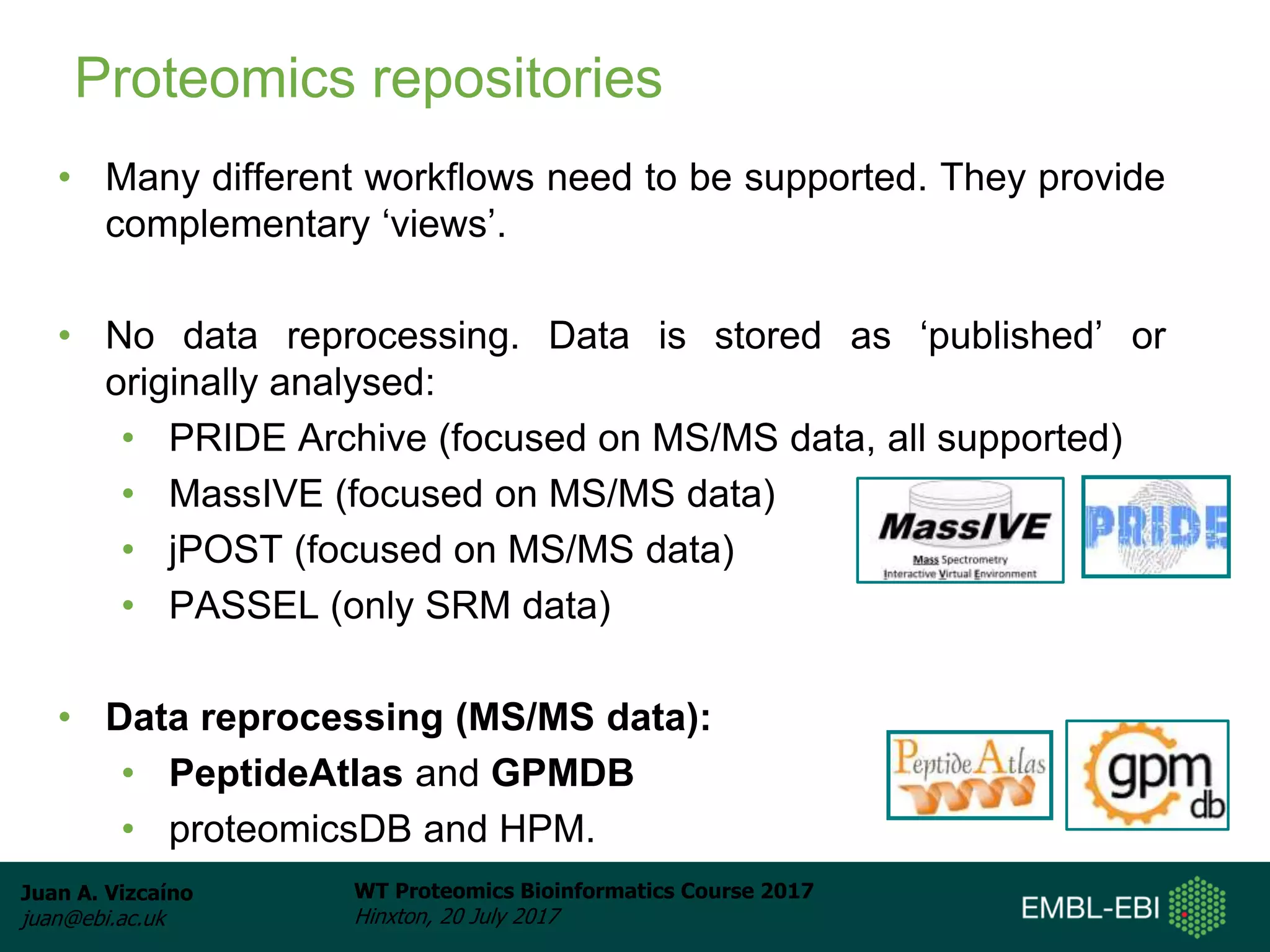
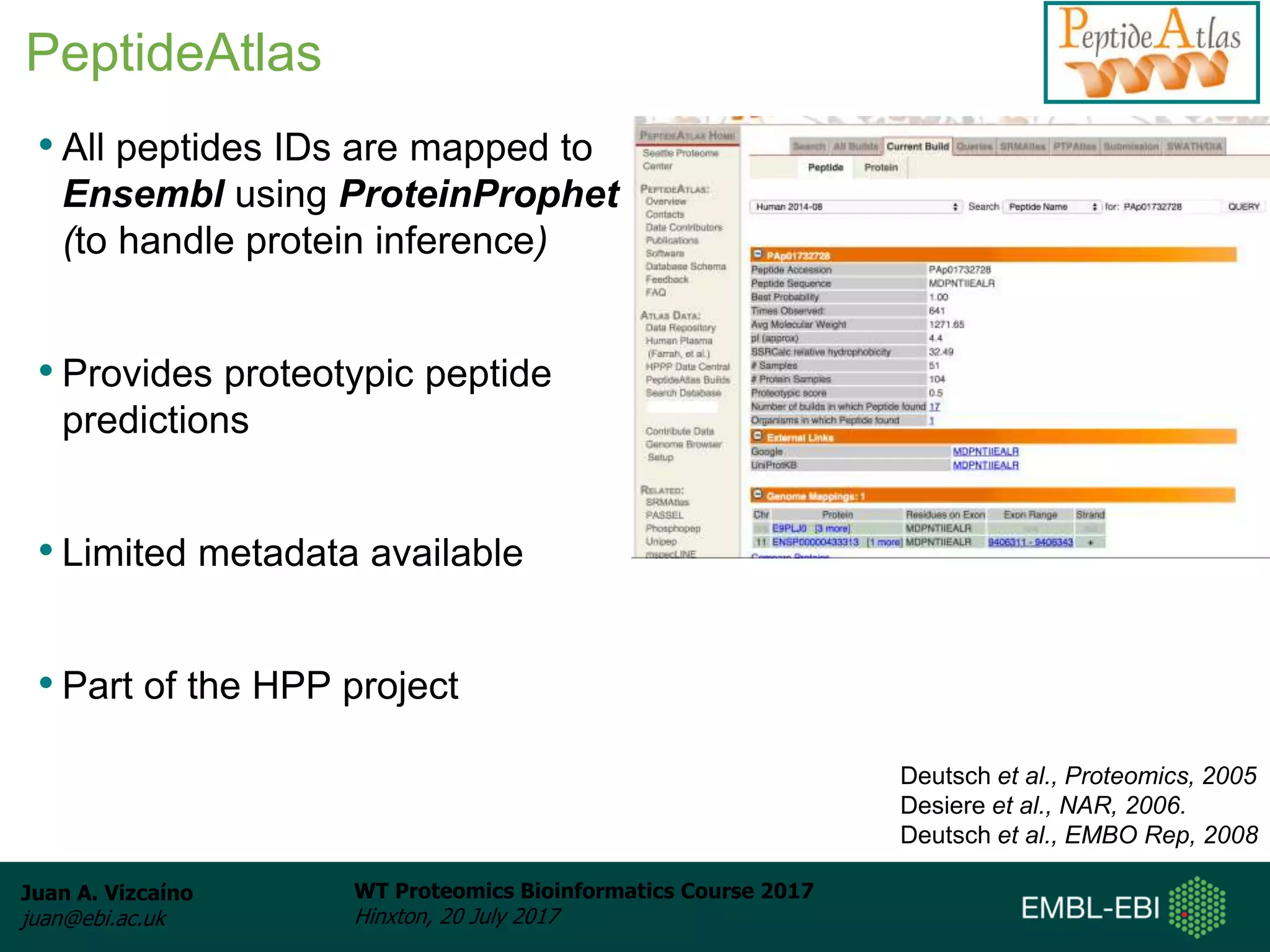
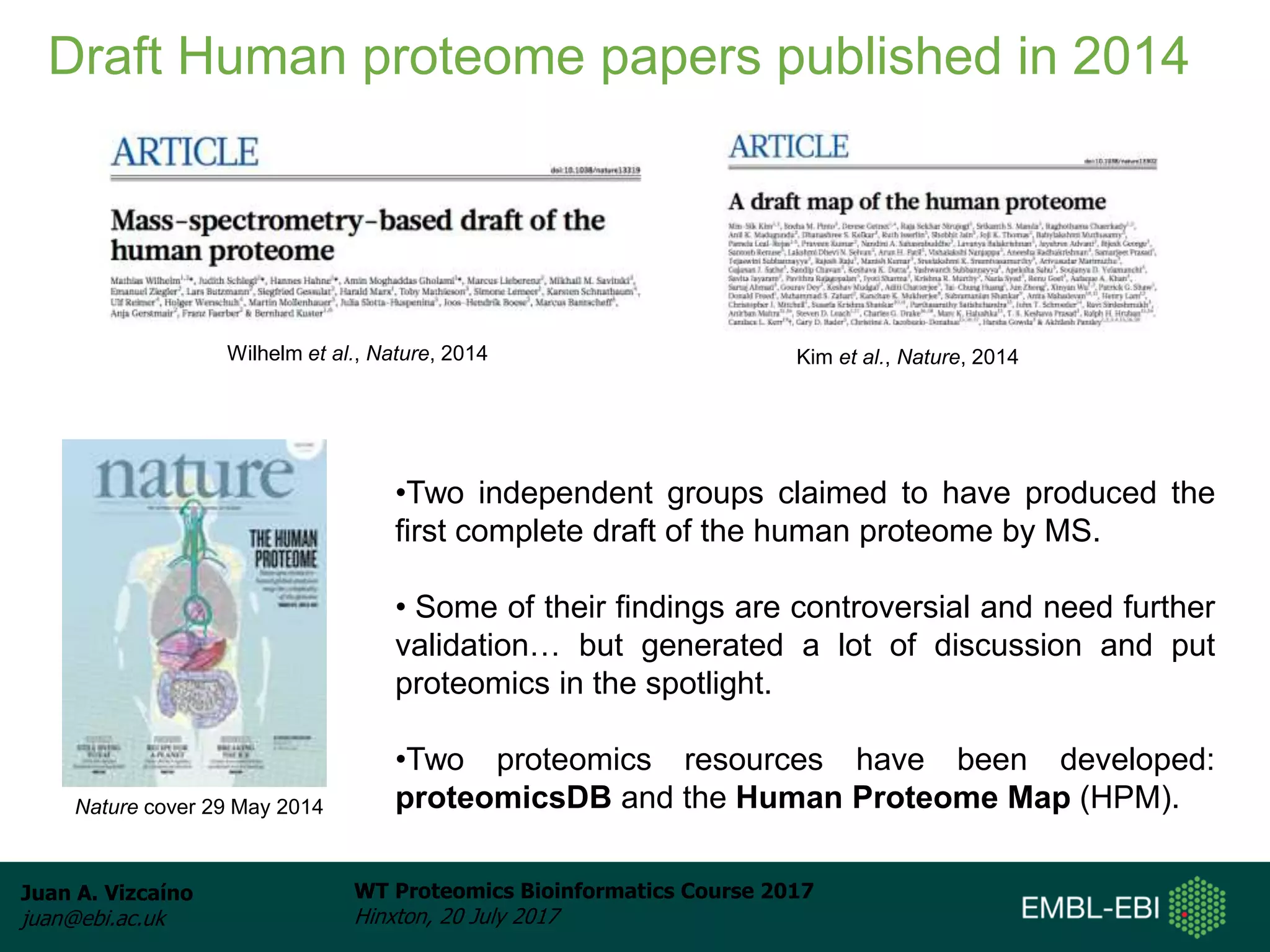
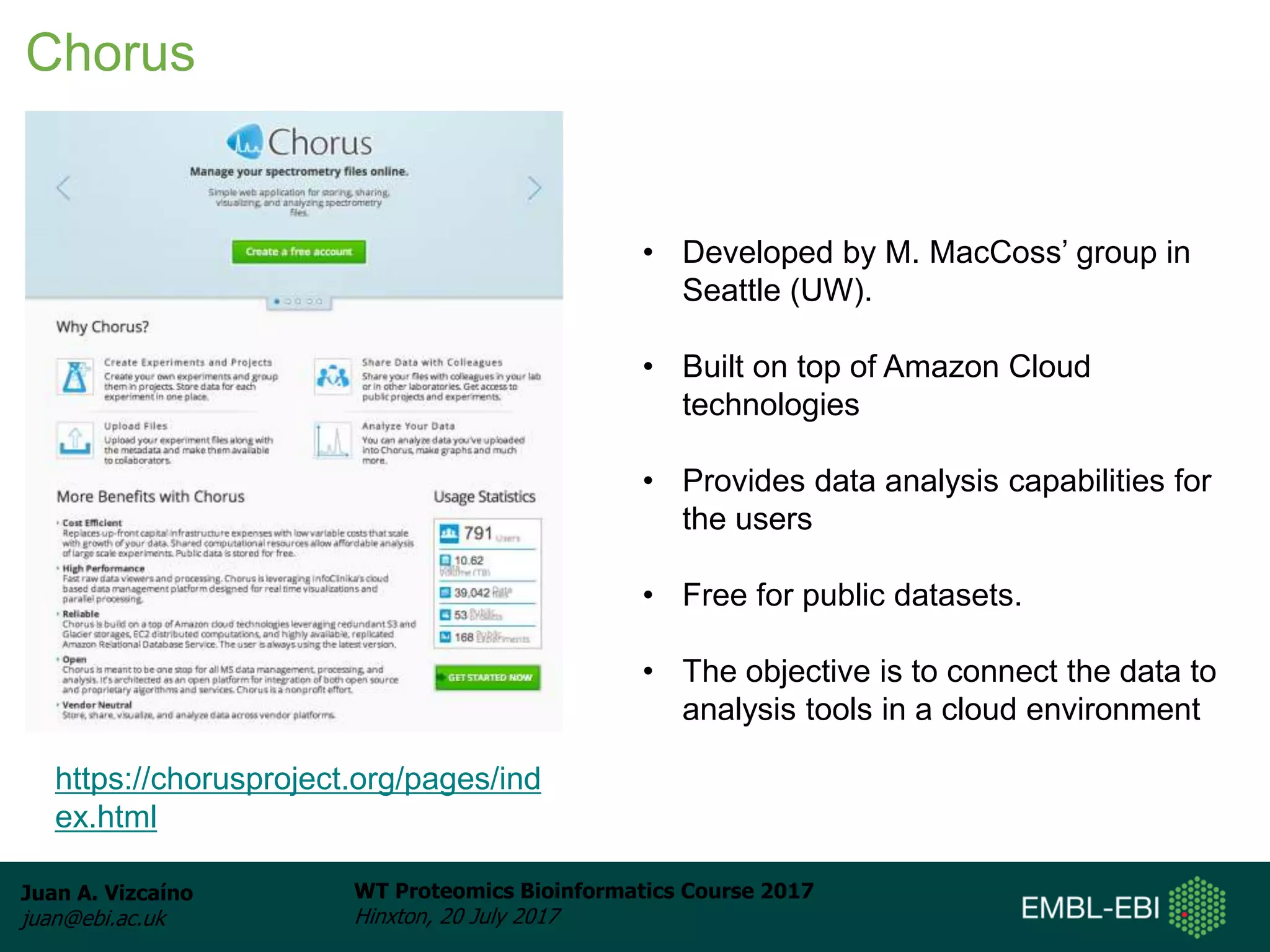
This document discusses proteomics repositories and data sharing in proteomics. It describes the types of information stored in MS proteomics repositories, including raw data, identification results, quantification, and metadata. It outlines several main repositories, distinguishing between those that do not reprocess data, like PRIDE and MassIVE, and those that do reprocess data through a standardized pipeline, like PeptideAtlas and GPMDB. It also discusses resources focused on drafts of the human proteome, such as proteomicsDB and the Human Proteome Map. Overall, the document provides an overview of existing proteomics repositories and issues around data sharing in the field.