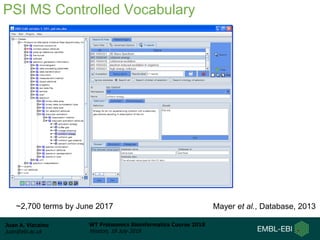
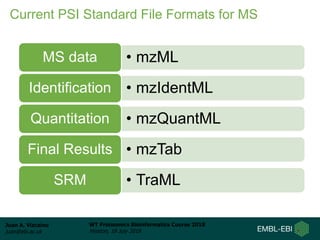
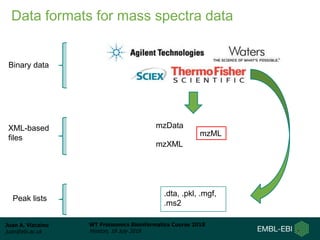
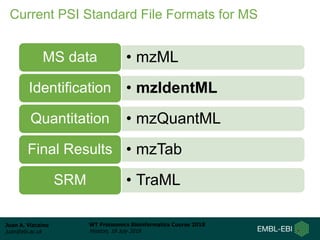
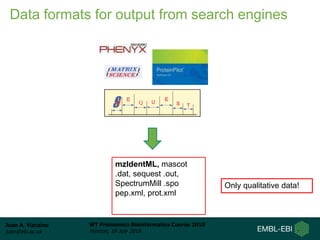
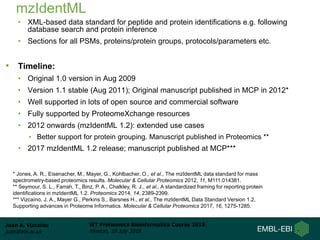
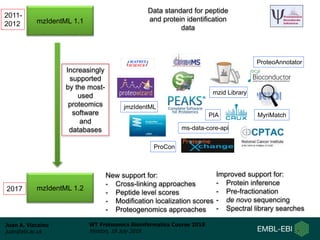
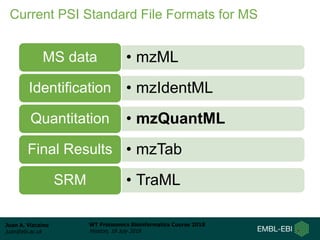
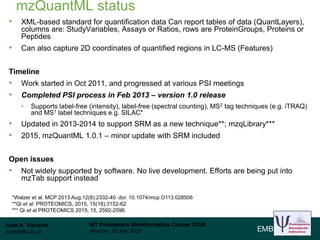
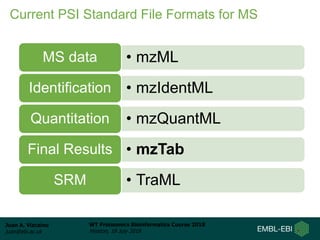
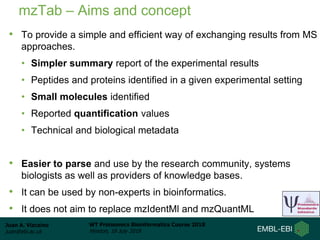
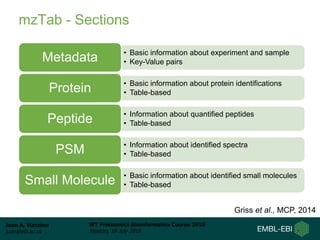
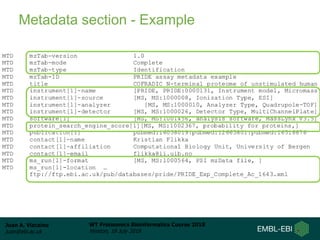
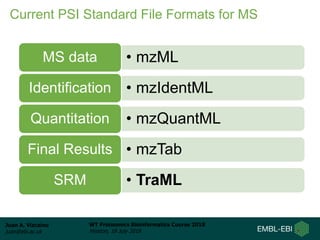
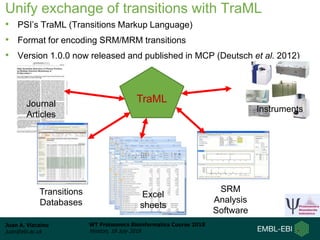
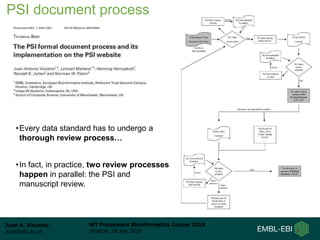
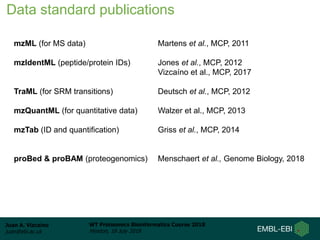
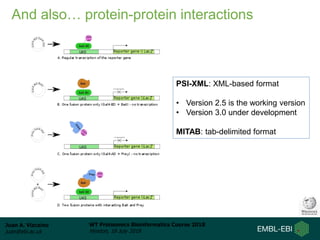
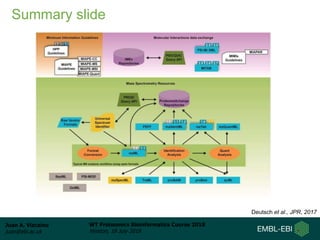
This document provides an overview of proteomics data standards developed by the Proteomics Standards Initiative (PSI). It discusses the need for data standards, describes existing PSI standards like mzML for mass spectrometry data, mzIdentML for identification data, and mzTab for final results. The document also provides background on the development and adoption of these standards over time to support the evolving needs of the proteomics community.