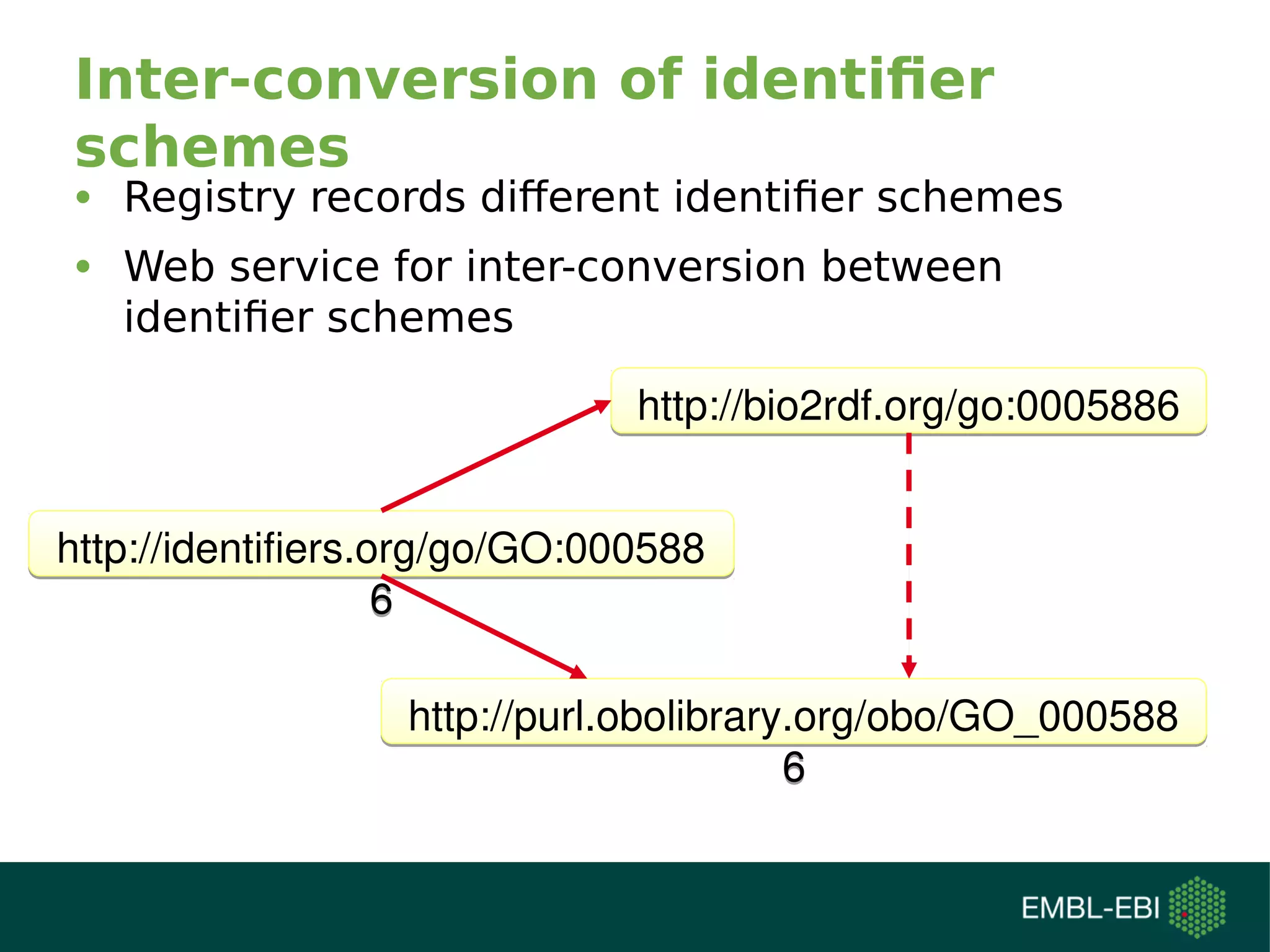
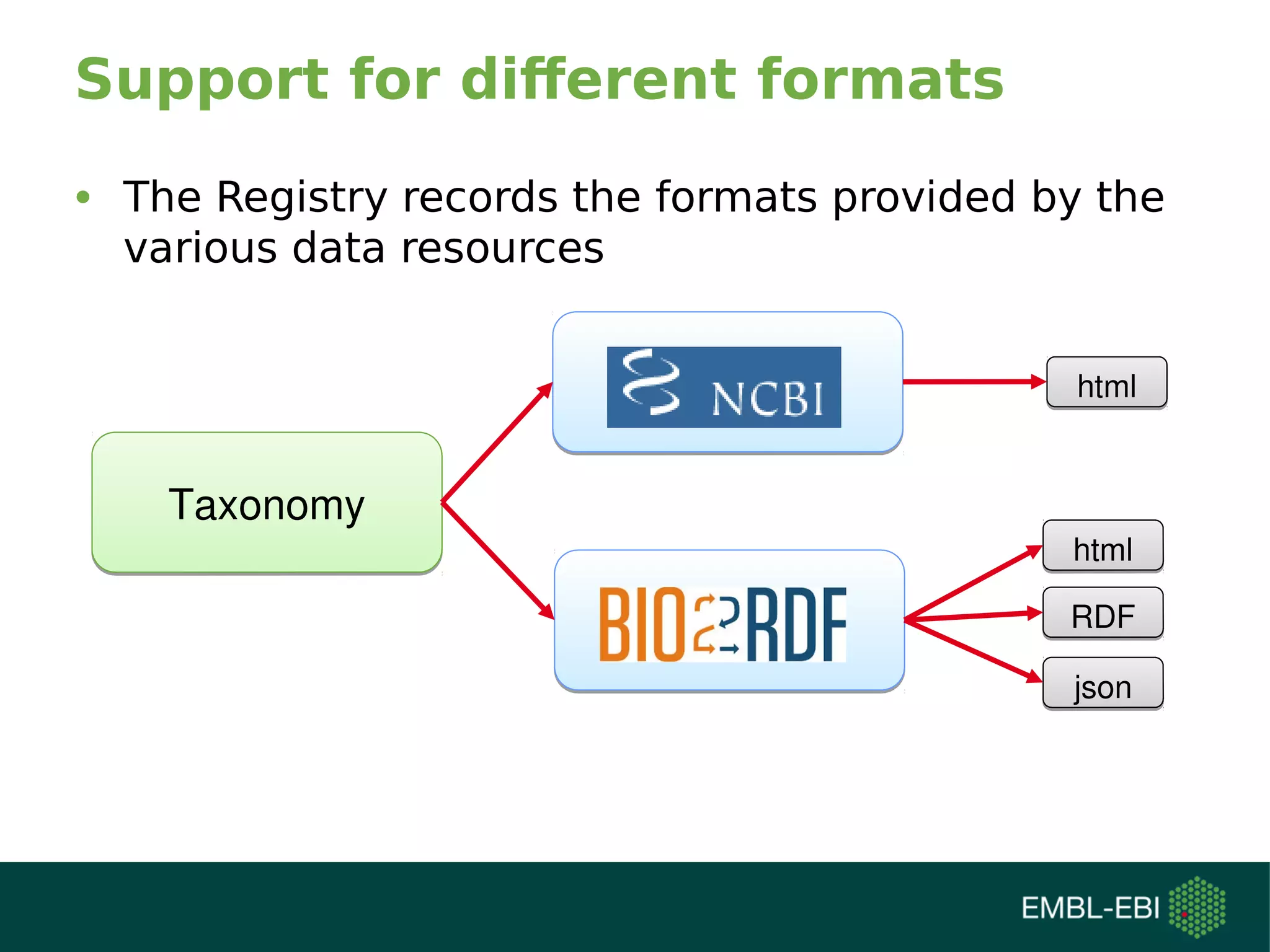
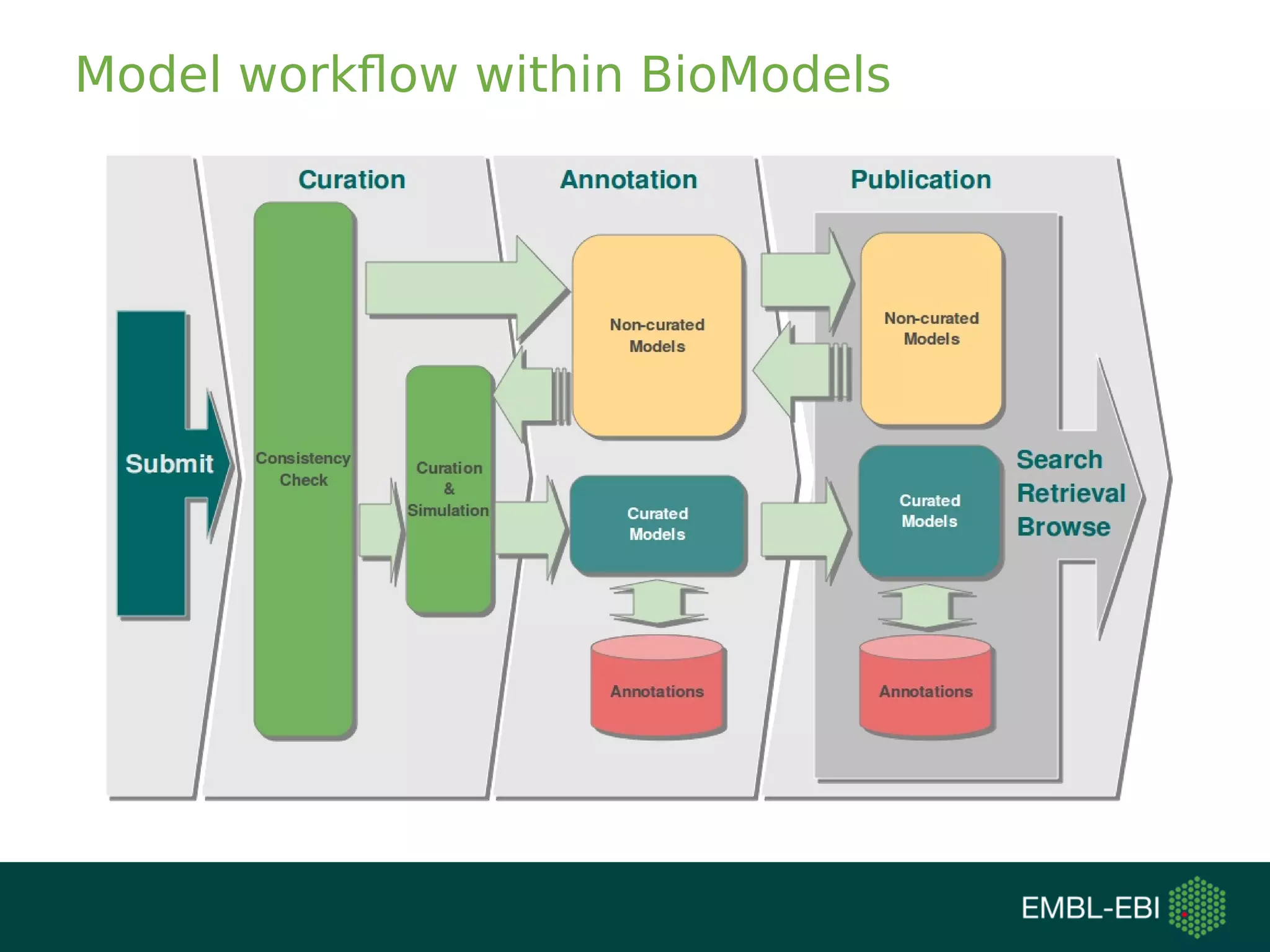
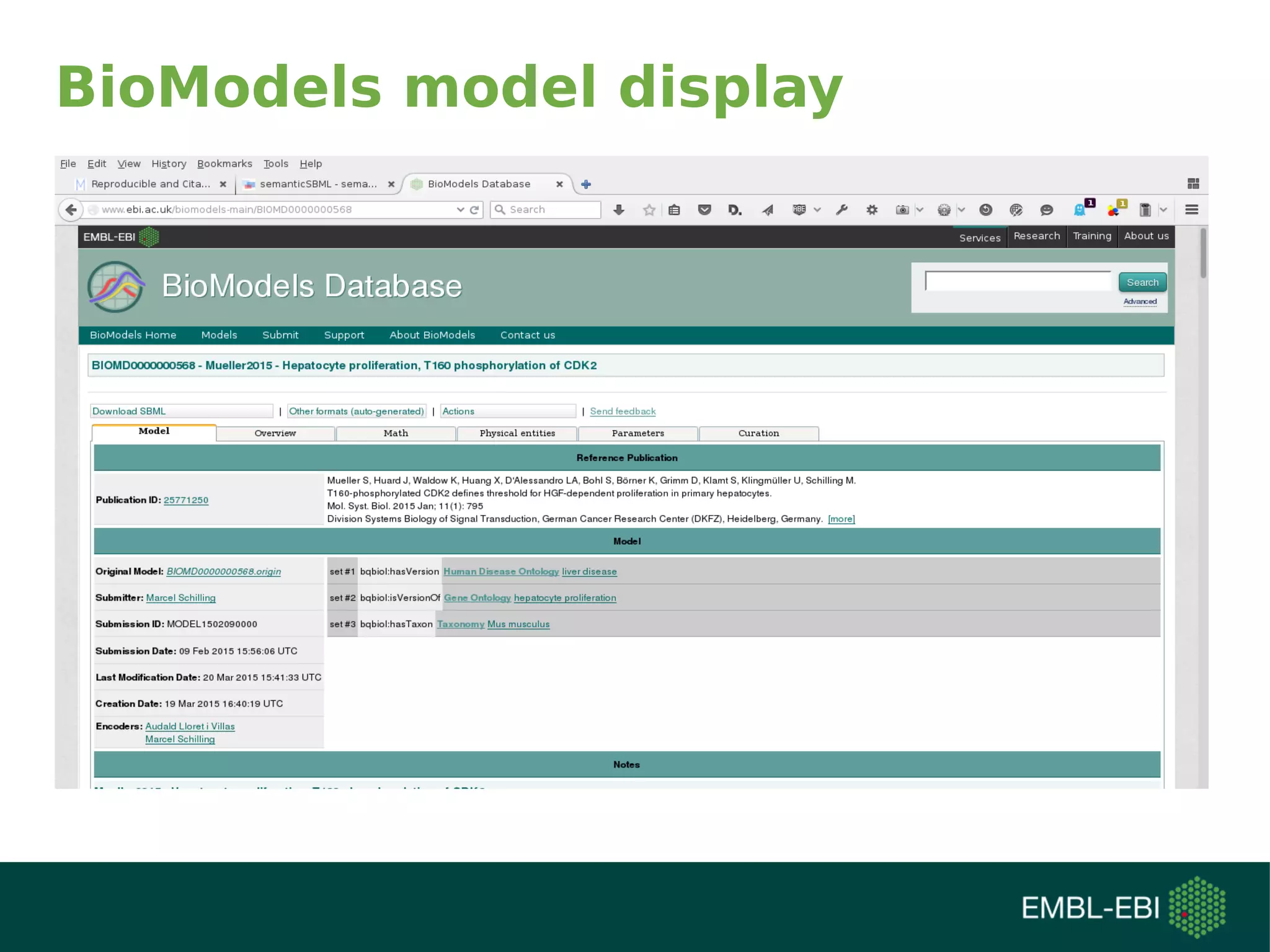
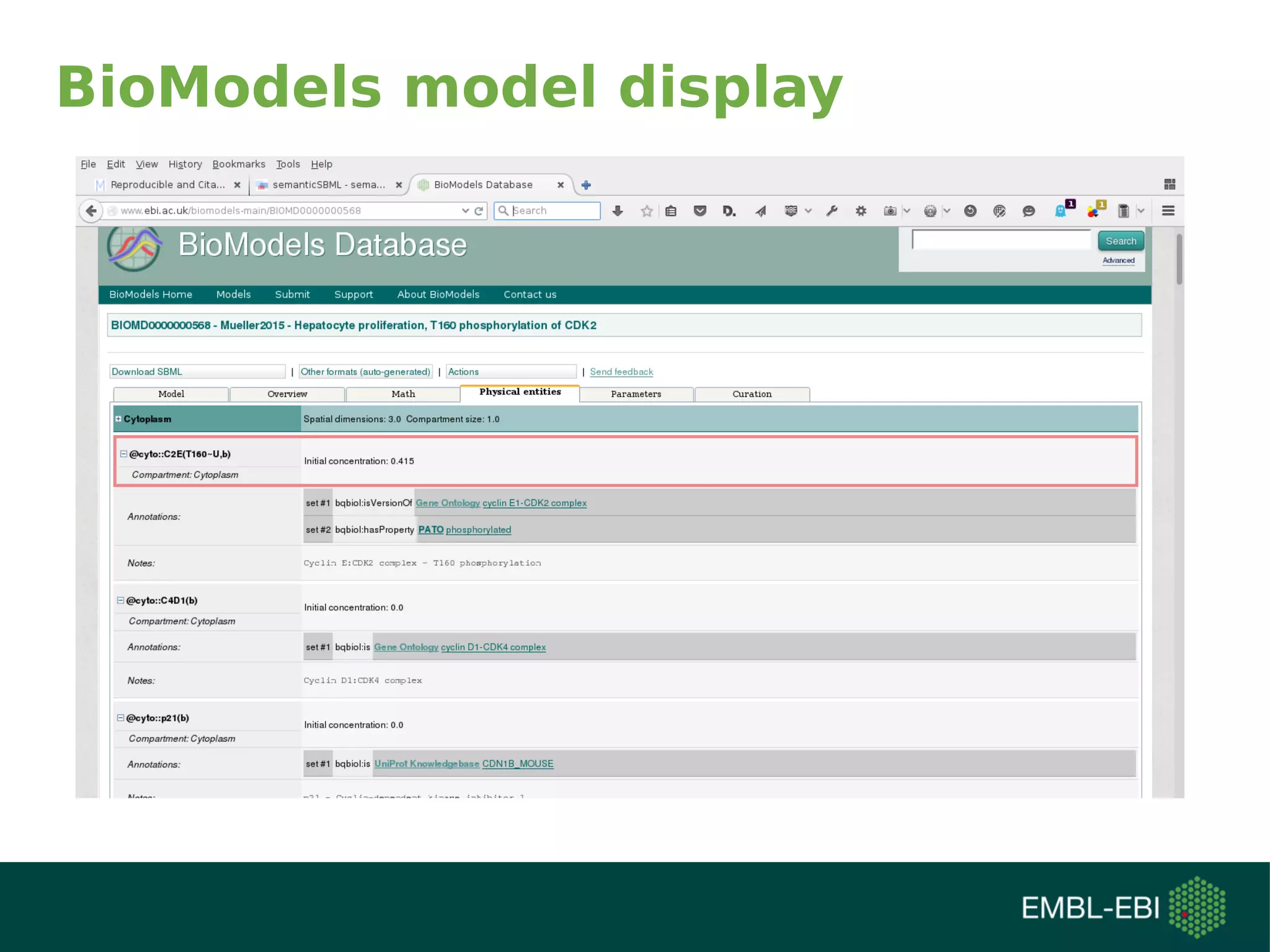
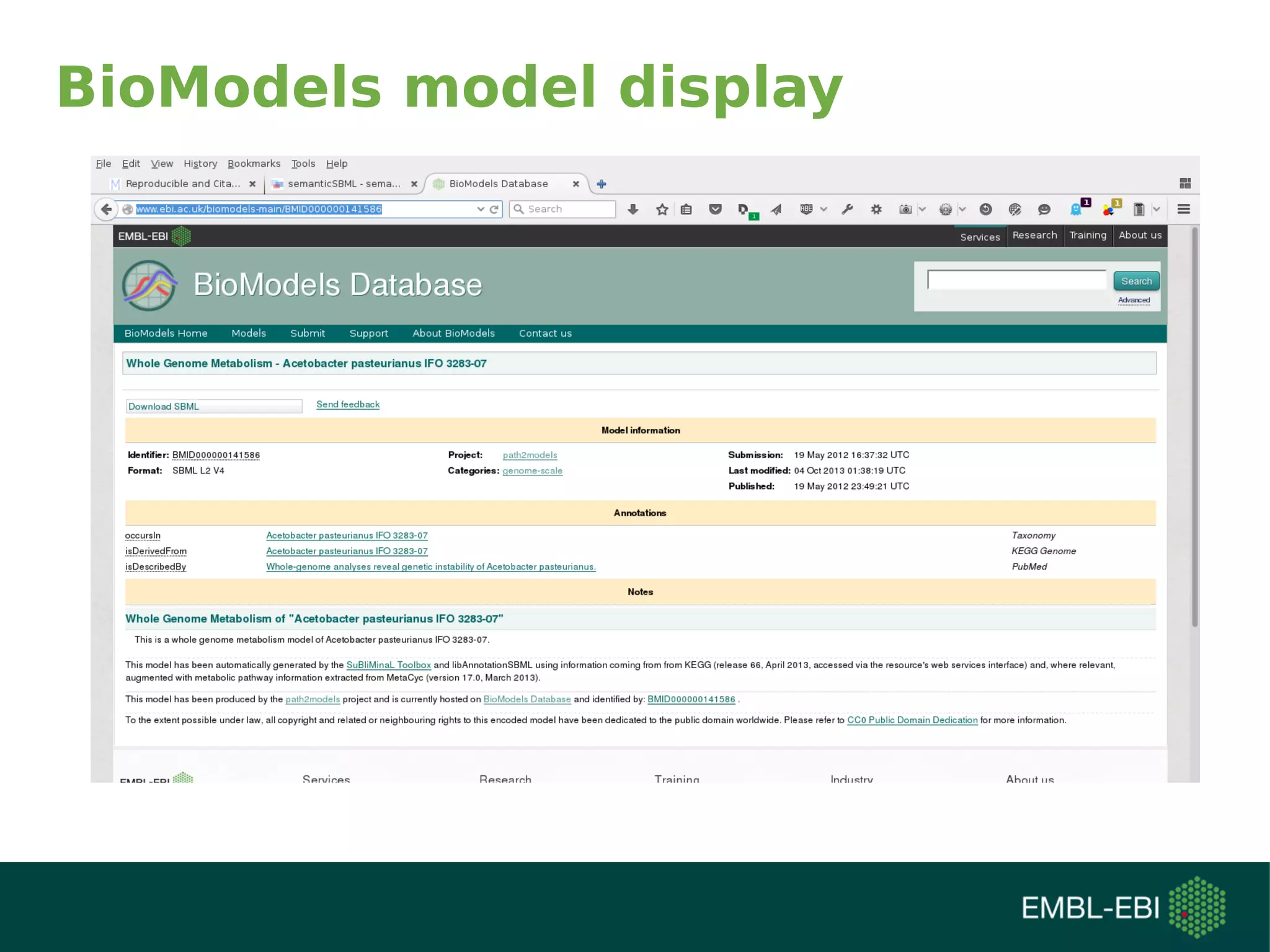
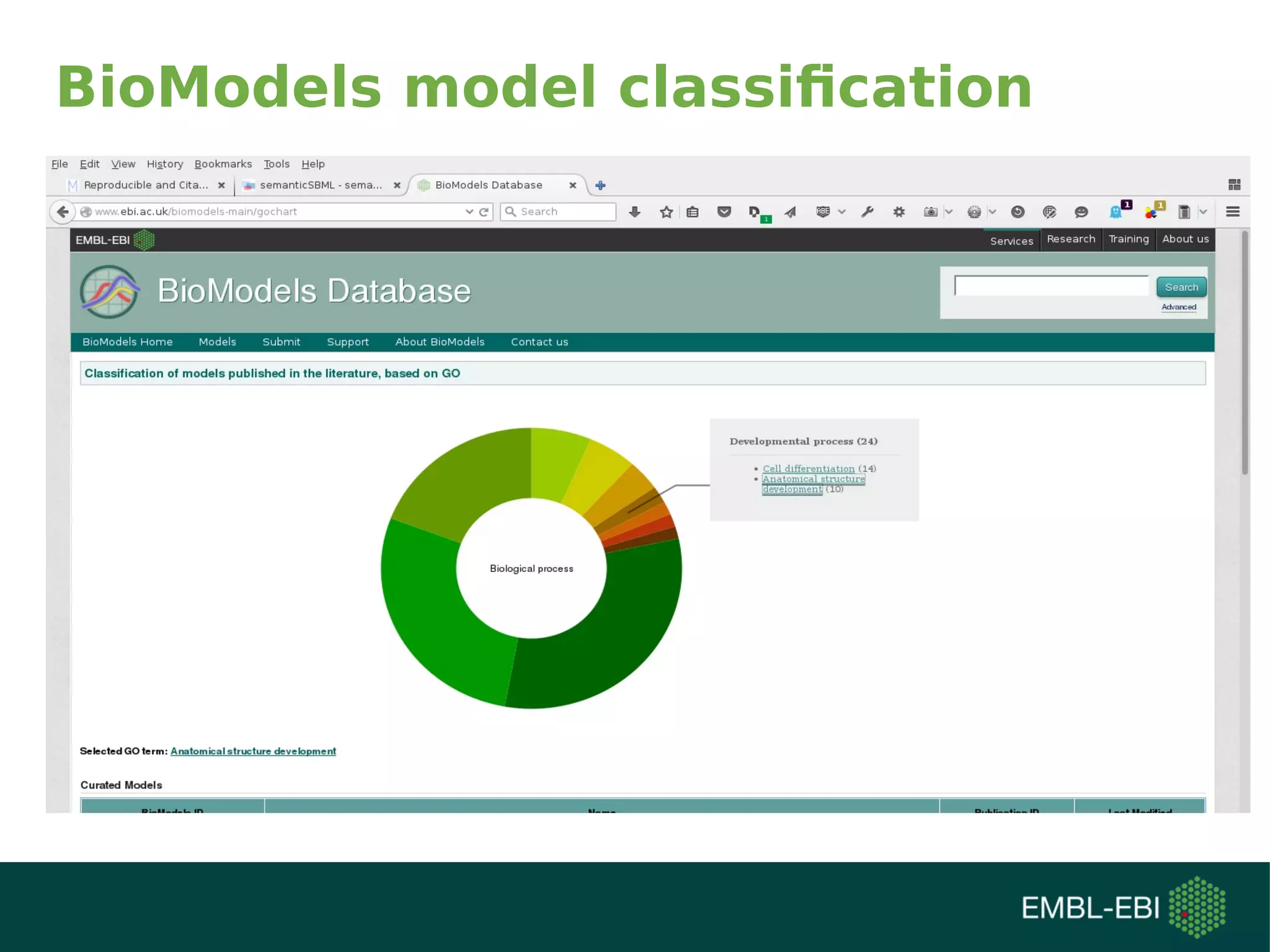
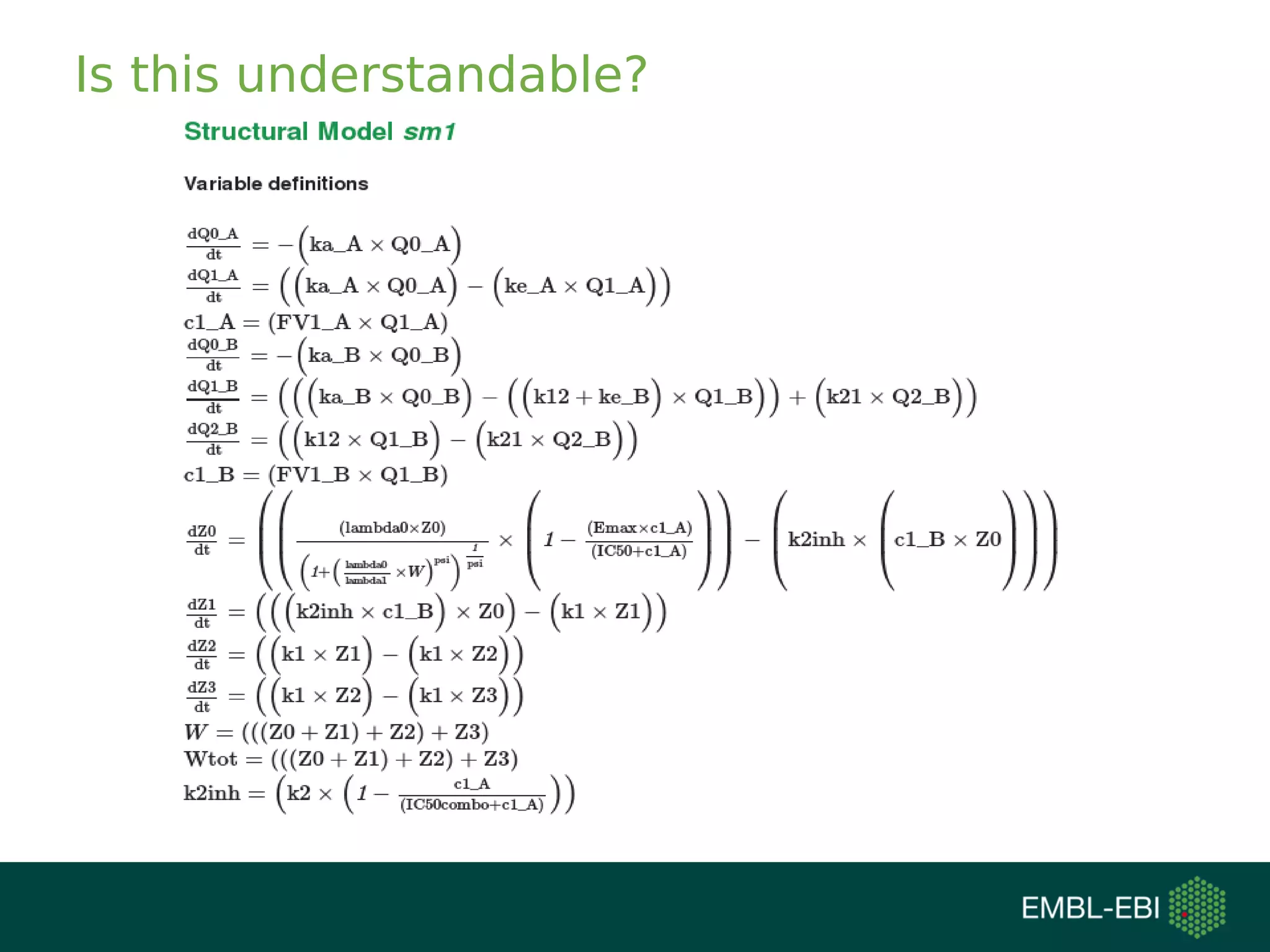
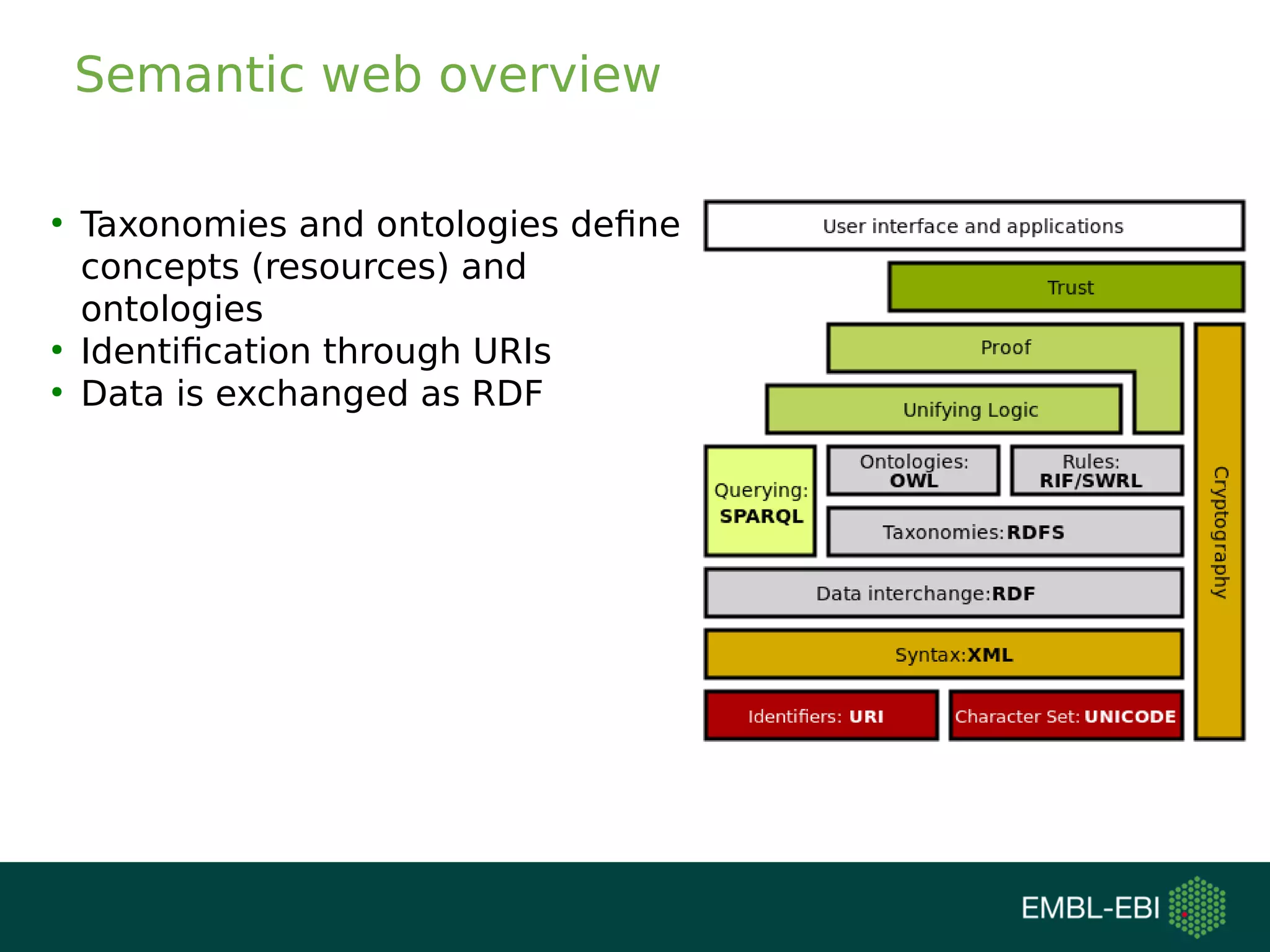
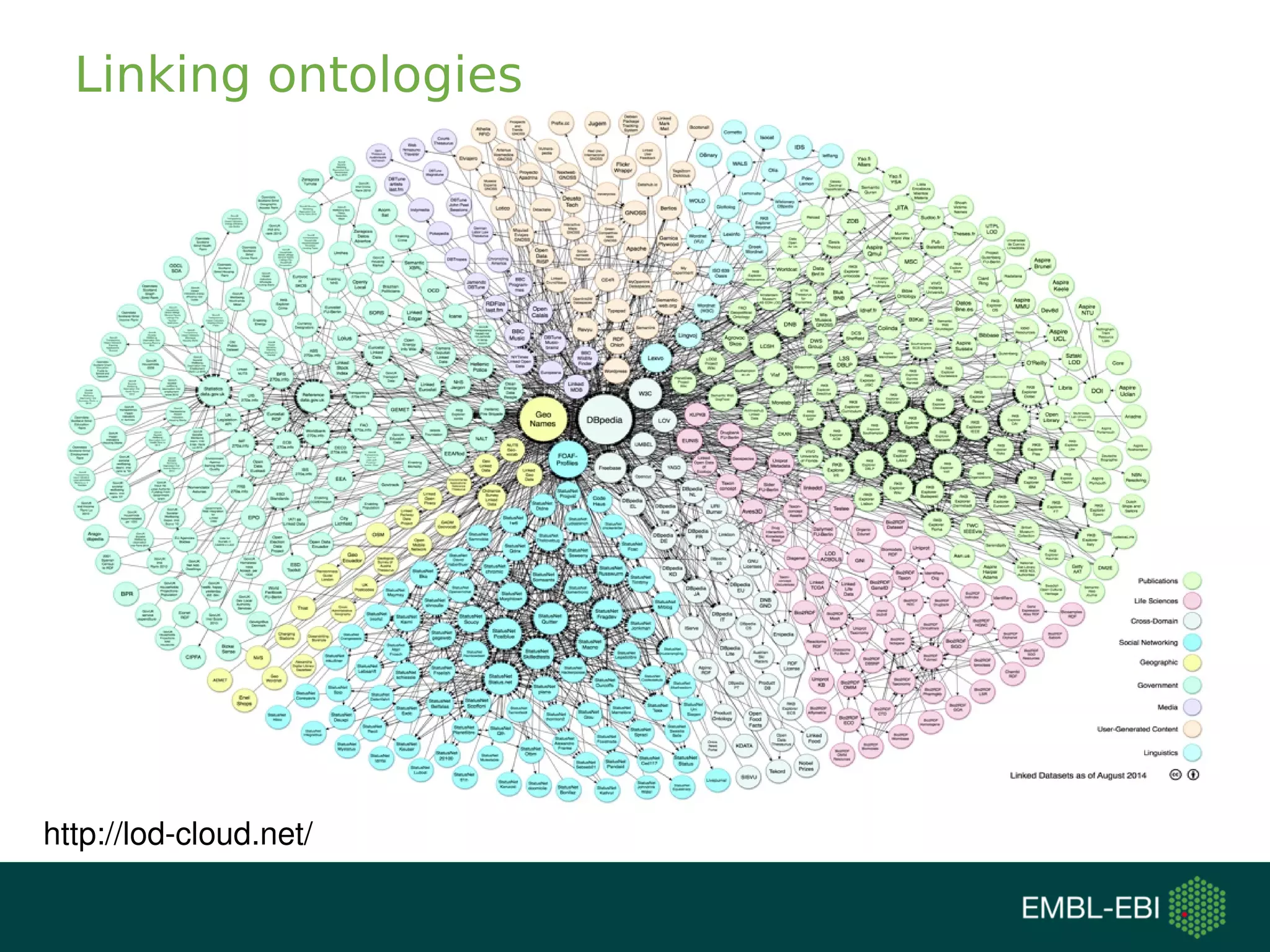
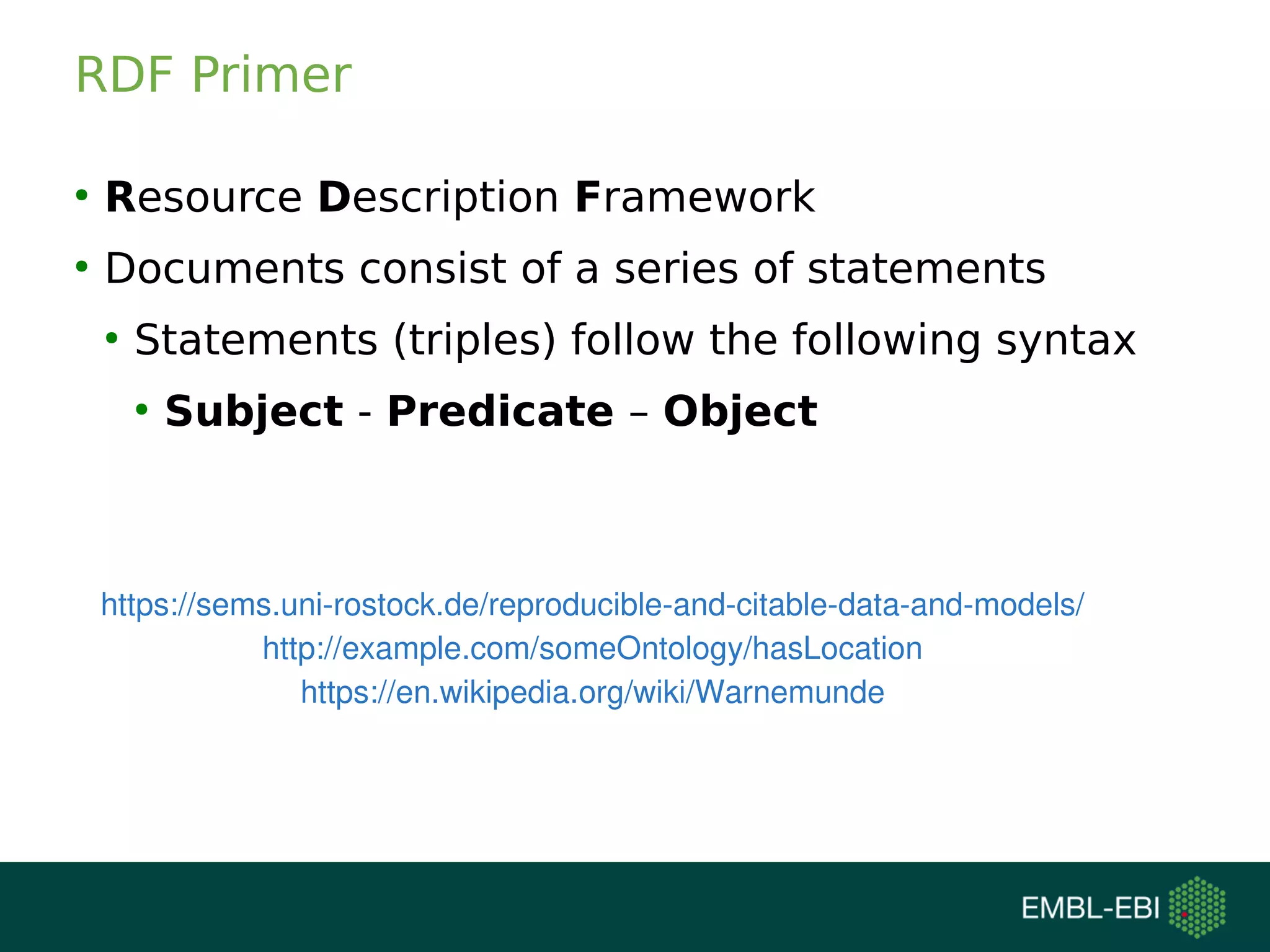
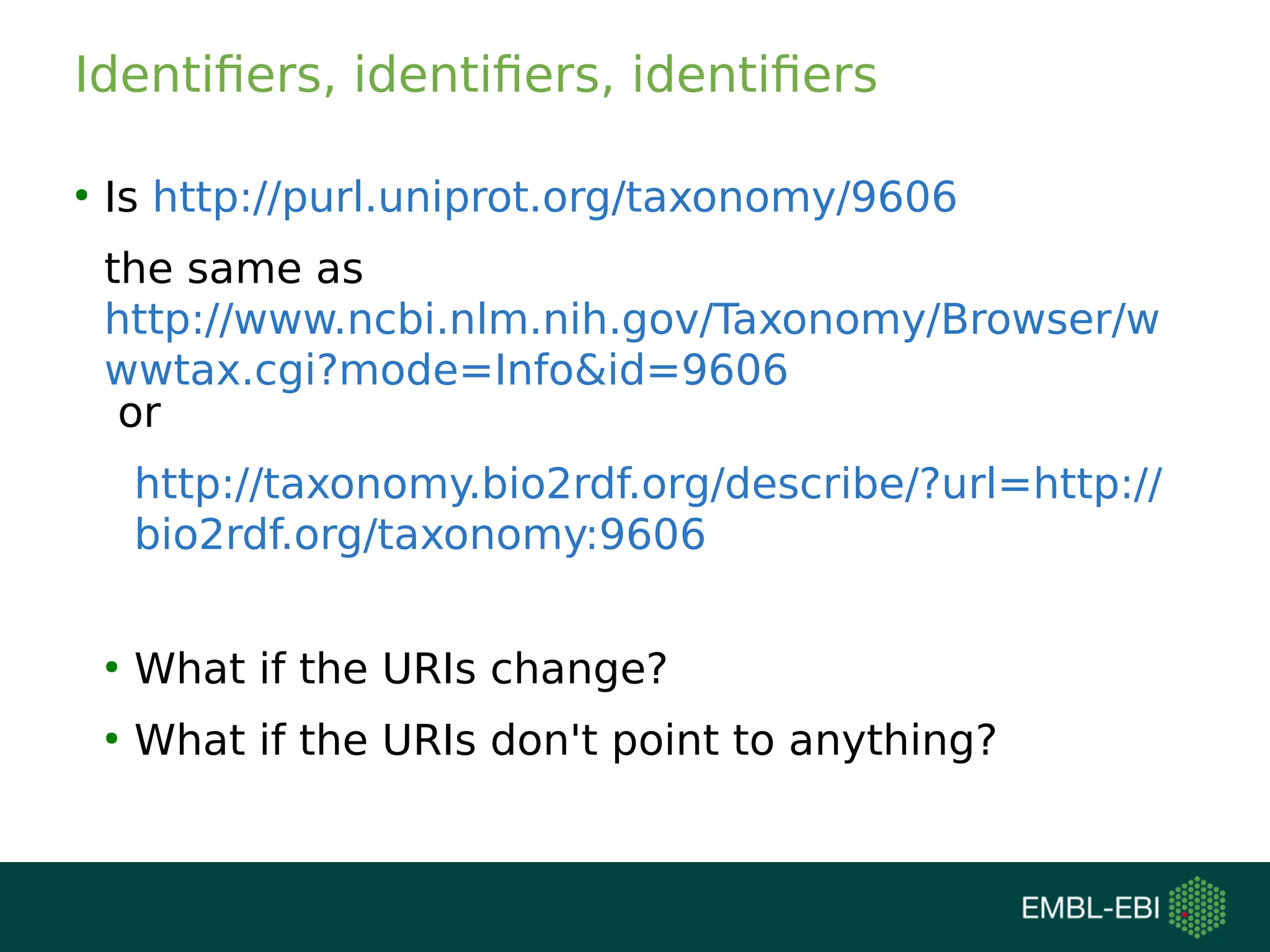
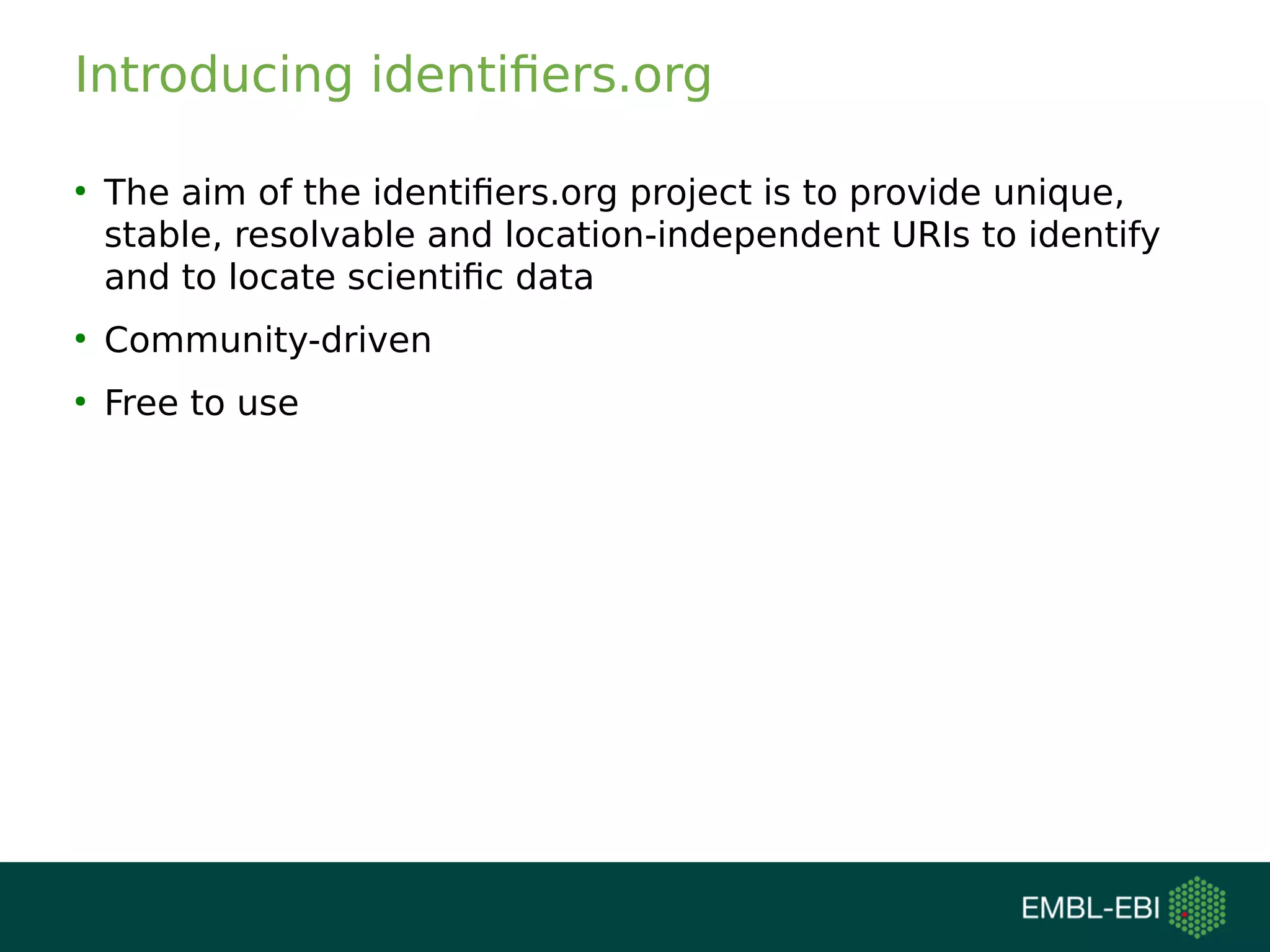
The document discusses challenges in finding reusable and reproducible modeling data, focusing on model identification and standardization. It highlights the evolution from Web 1.0 to Web 3.0, emphasizing the importance of semantic frameworks and unique identifiers for scientific data. The Identifiers.org project is introduced as a solution for providing stable and resolvable URIs for scientific data, alongside discussions on model curation and annotation.






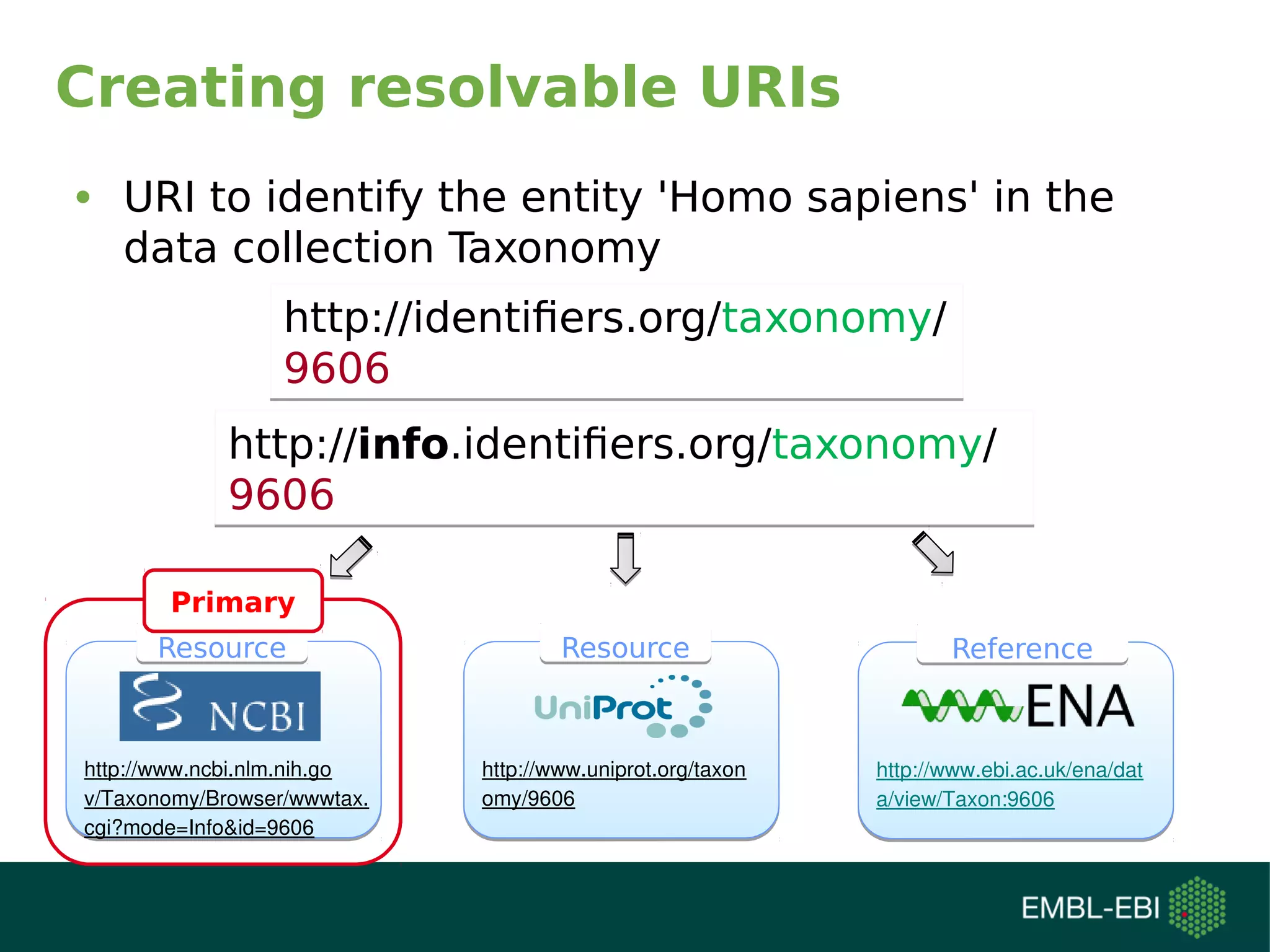
![Creating unique URIs
• Homo sapiens in Taxonomy
(9606)
http://identifiers.org/taxonomy/9606http://identifiers.org/taxonomy/9606
[Data
collection]
[Entity
identifier]](https://image.slidesharecdn.com/capturingcontext-150924135341-lva1-app6892/75/Capturing-the-context-one-small-ish-step-for-modellers-one-giant-leap-for-mankind-16-2048.jpg)