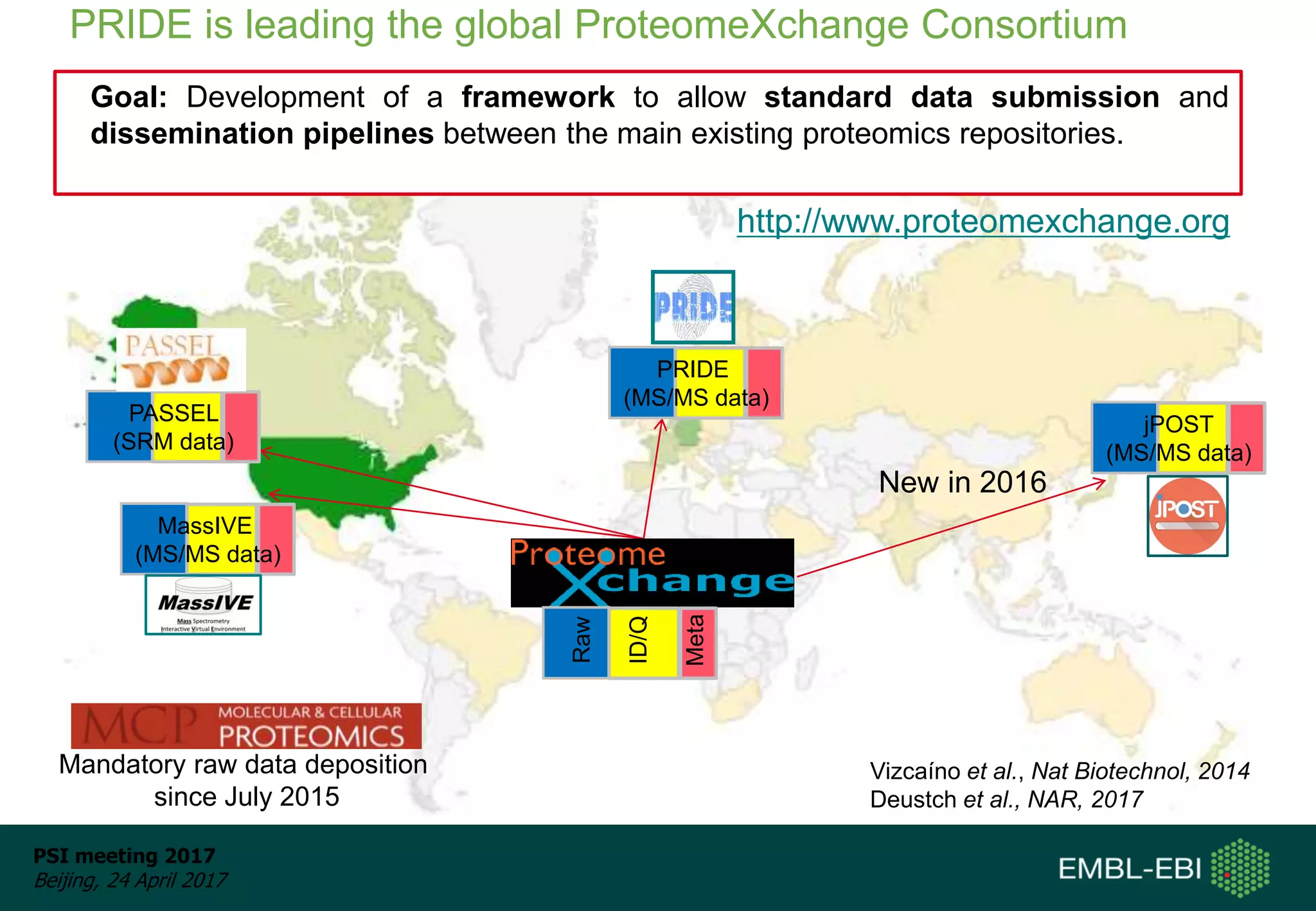
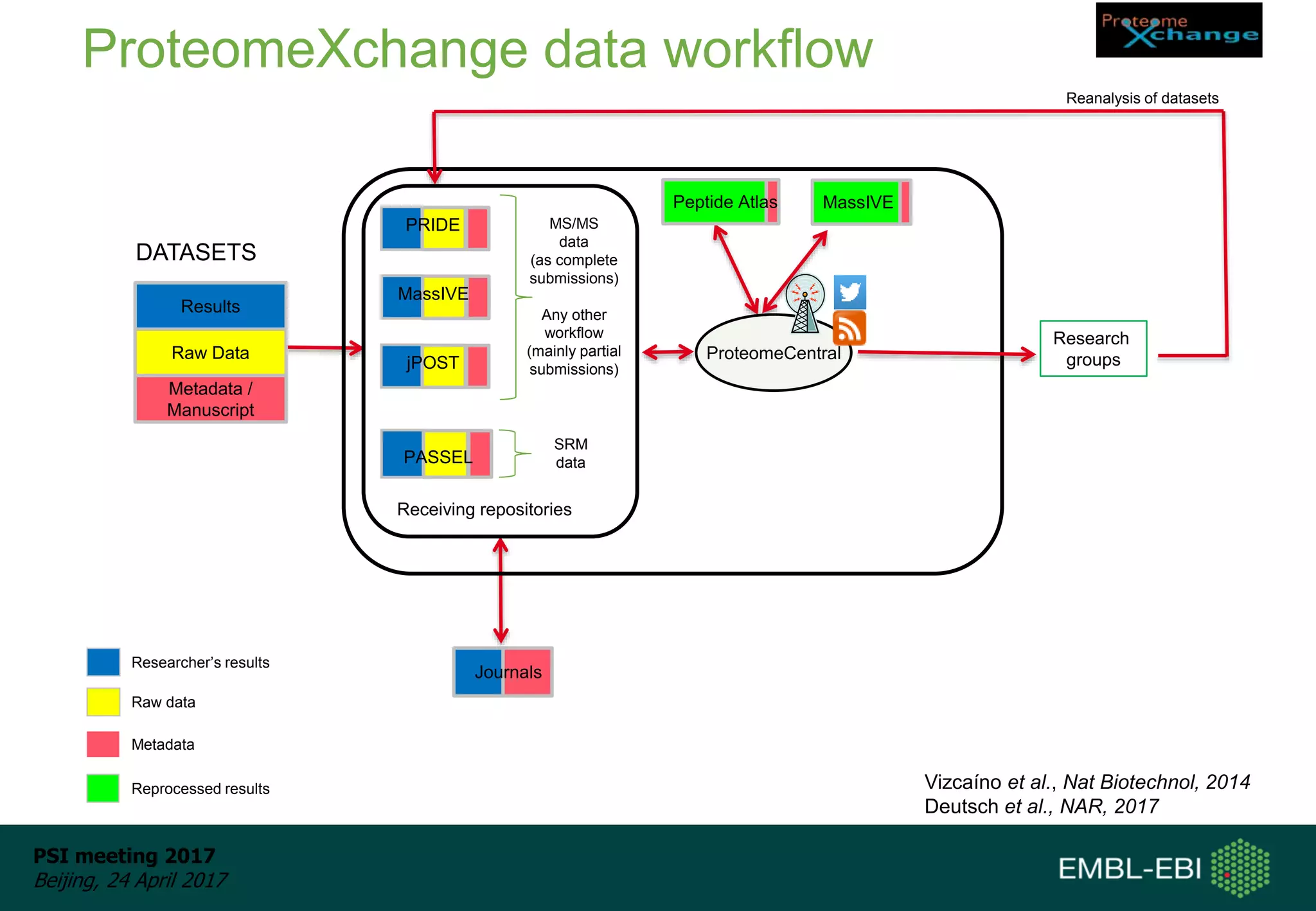
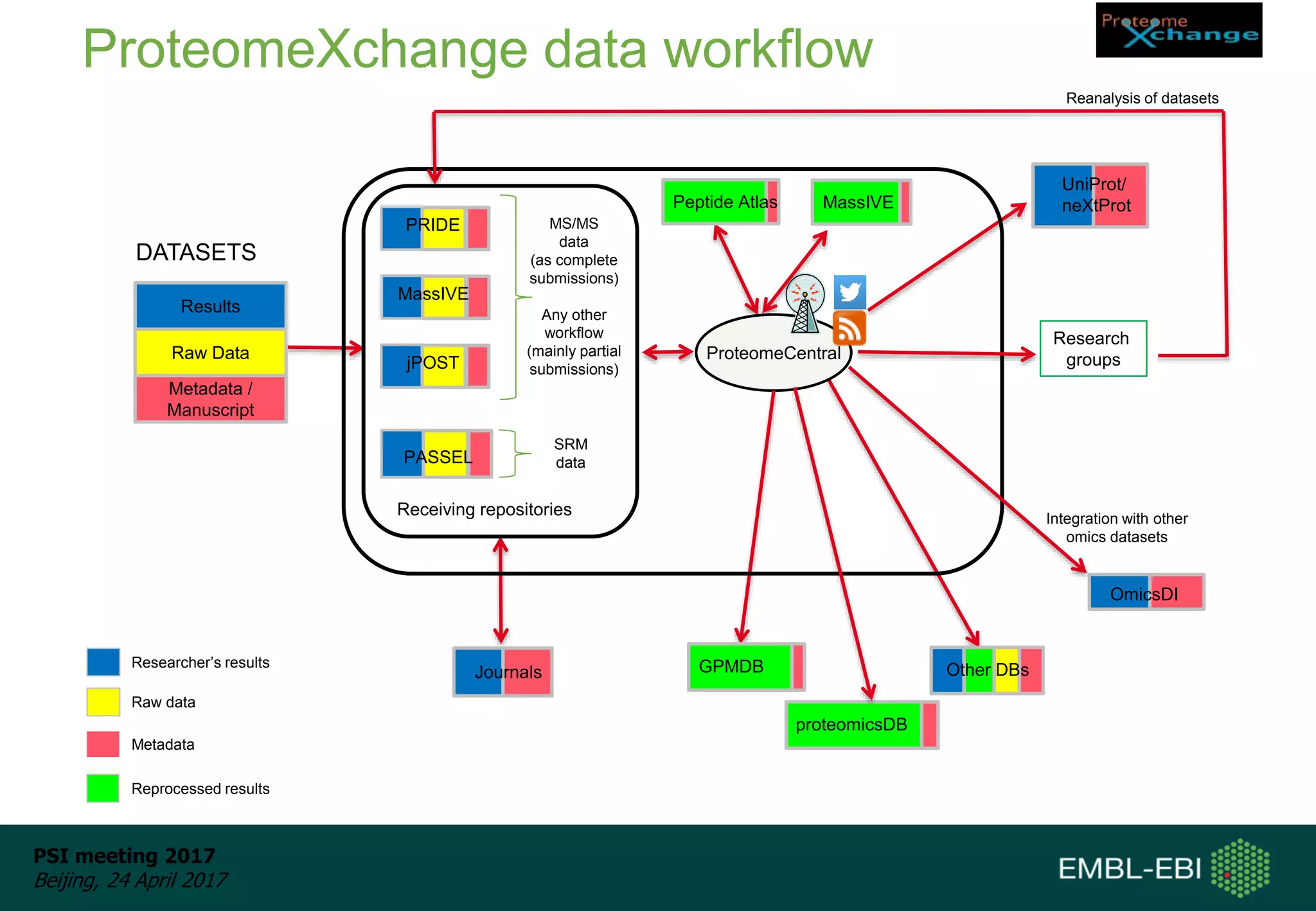
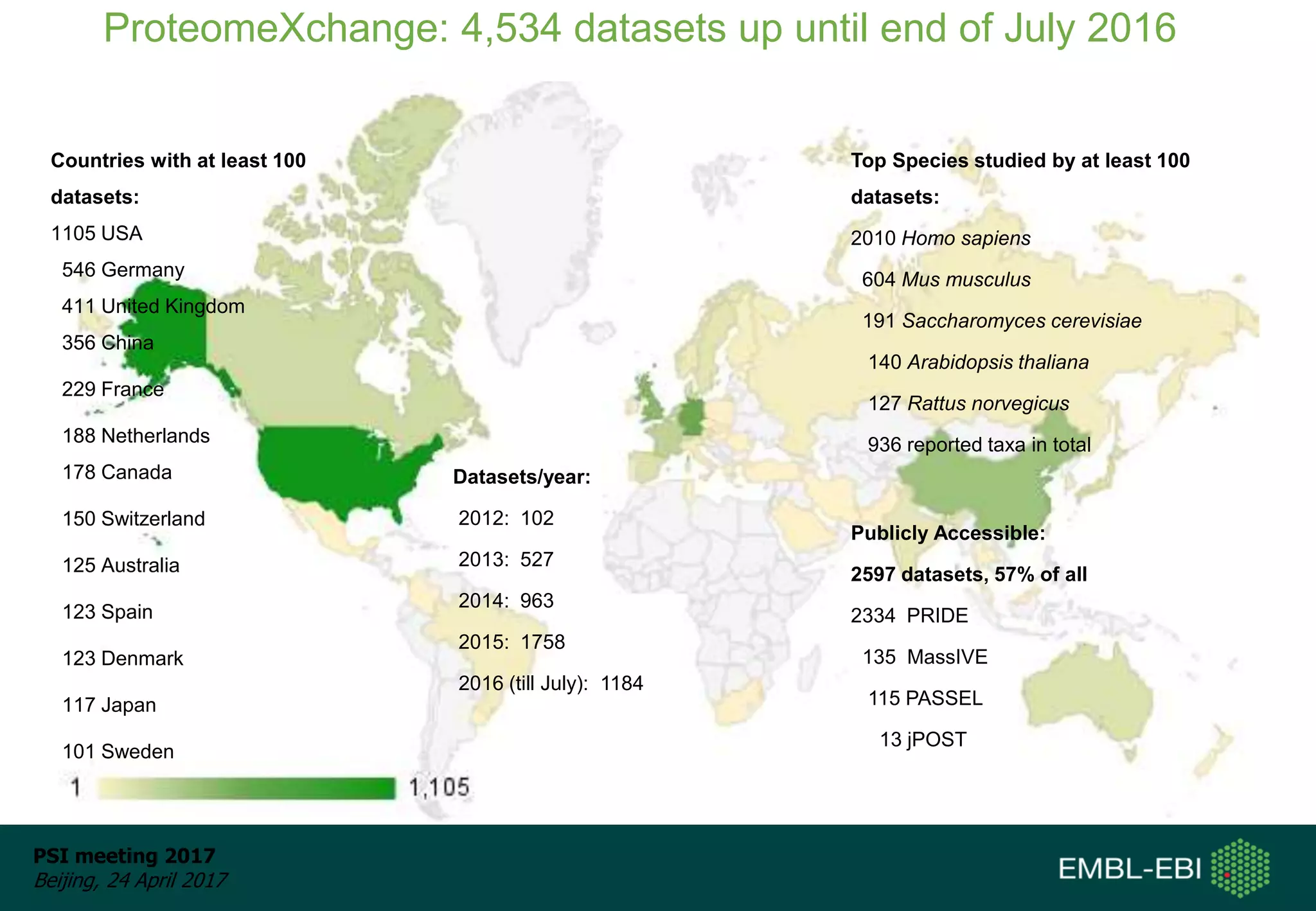
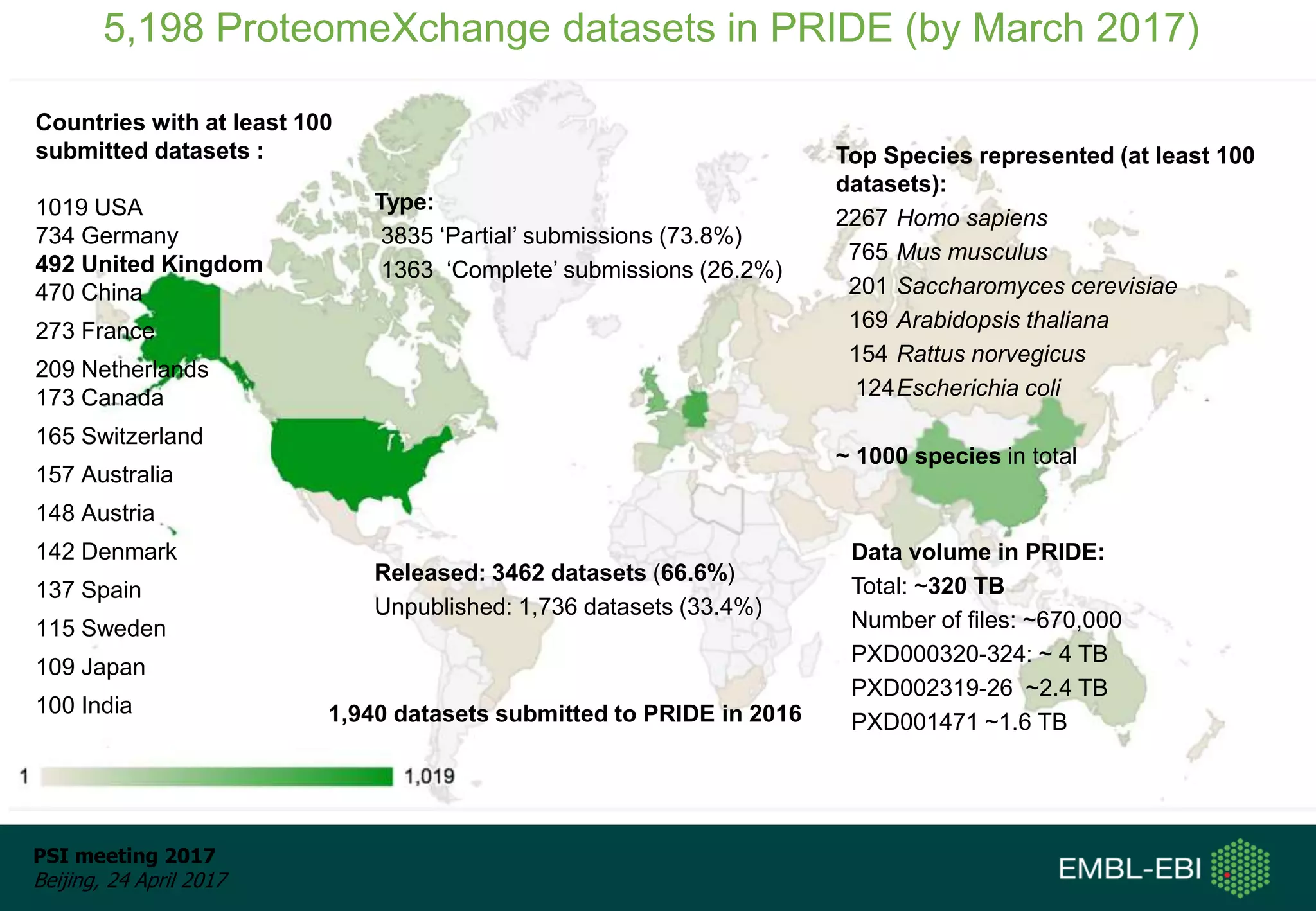
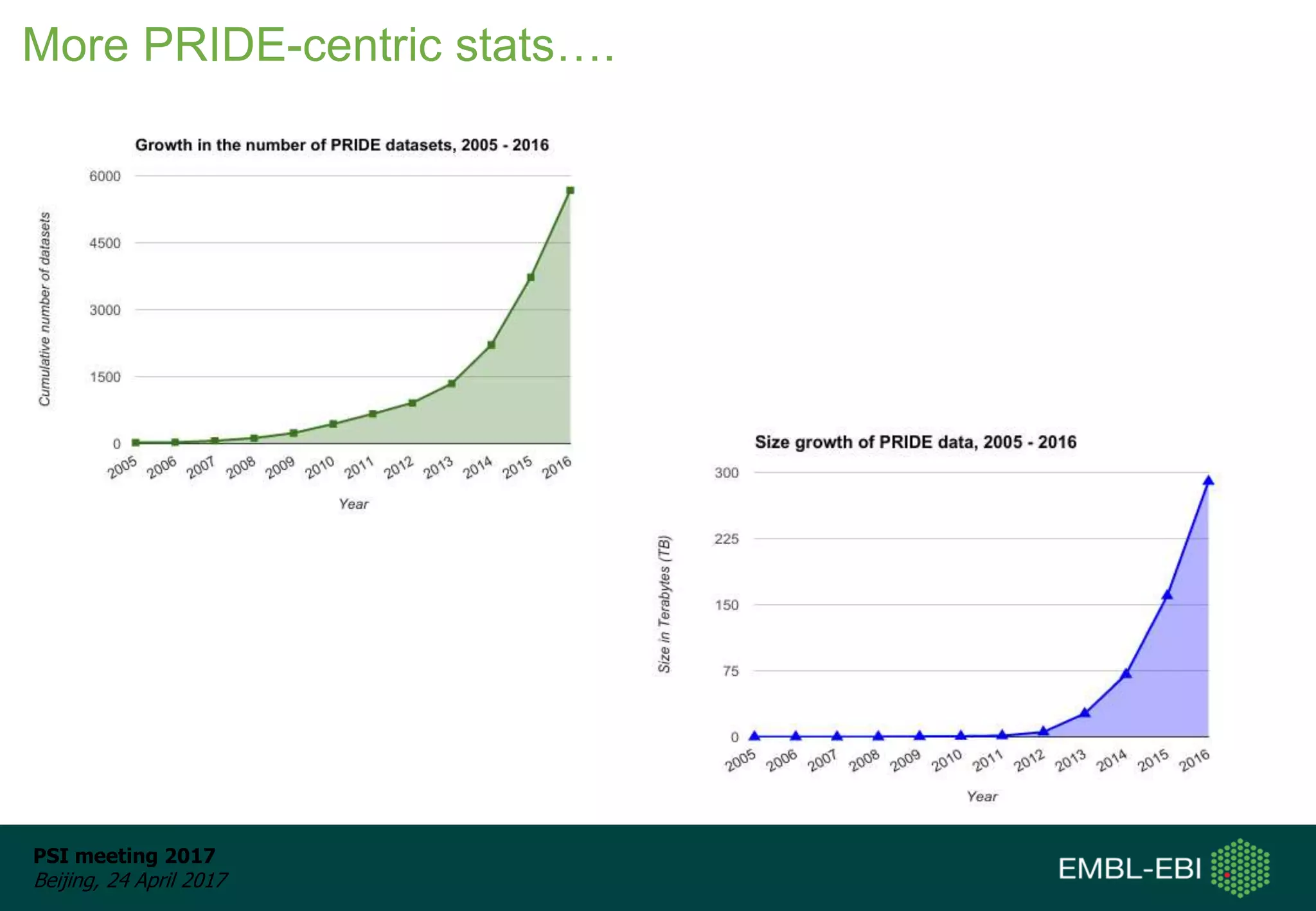
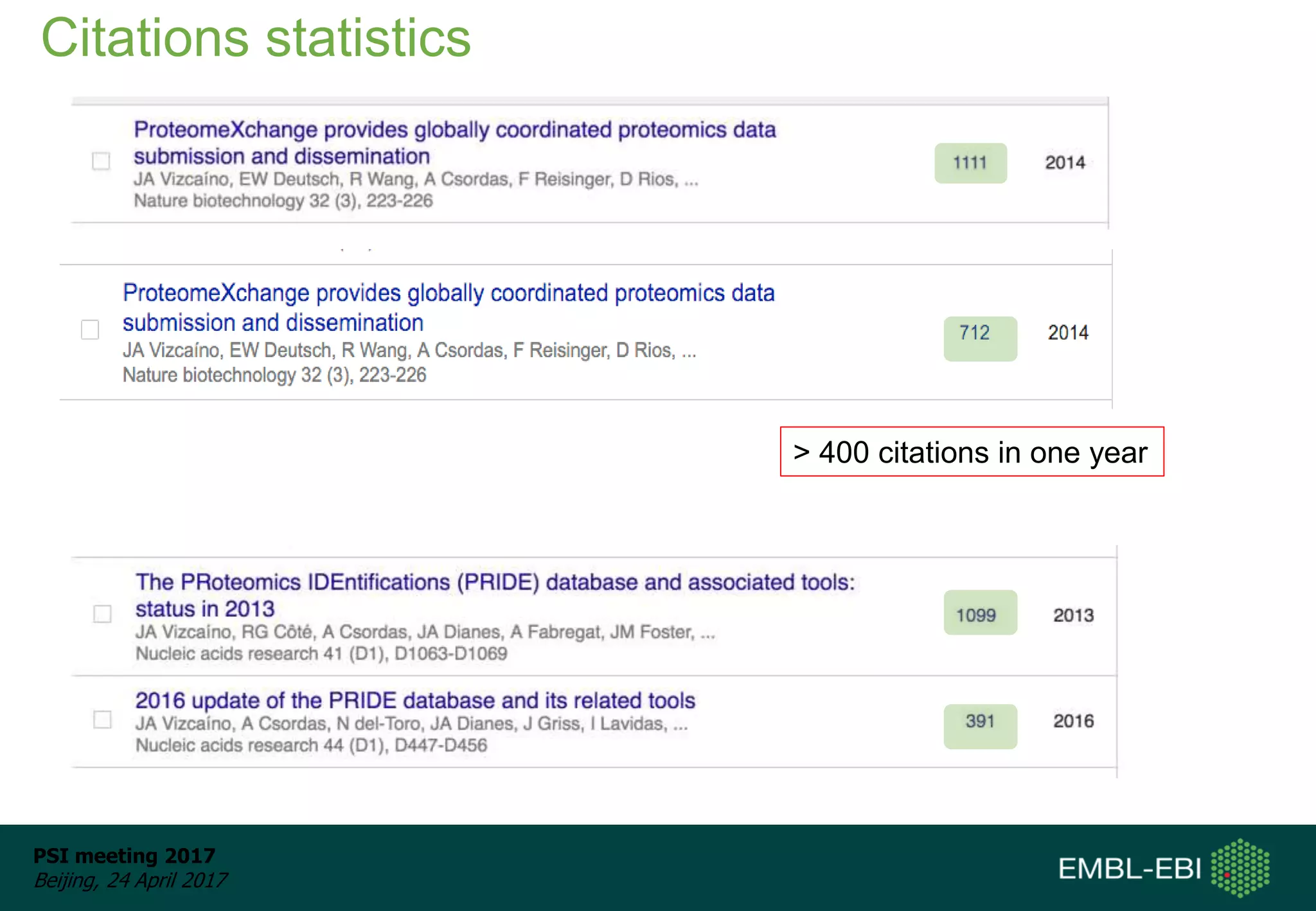
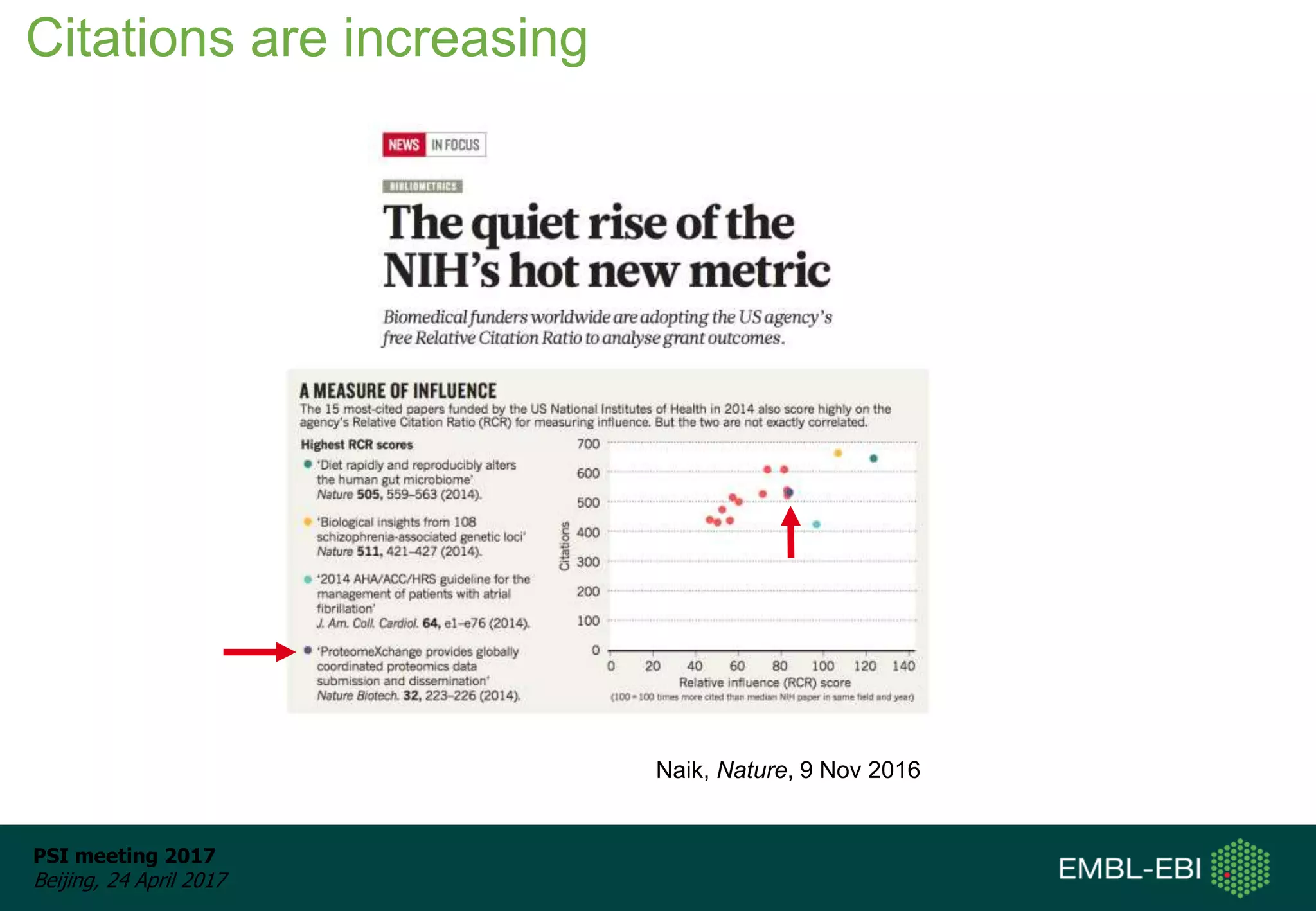
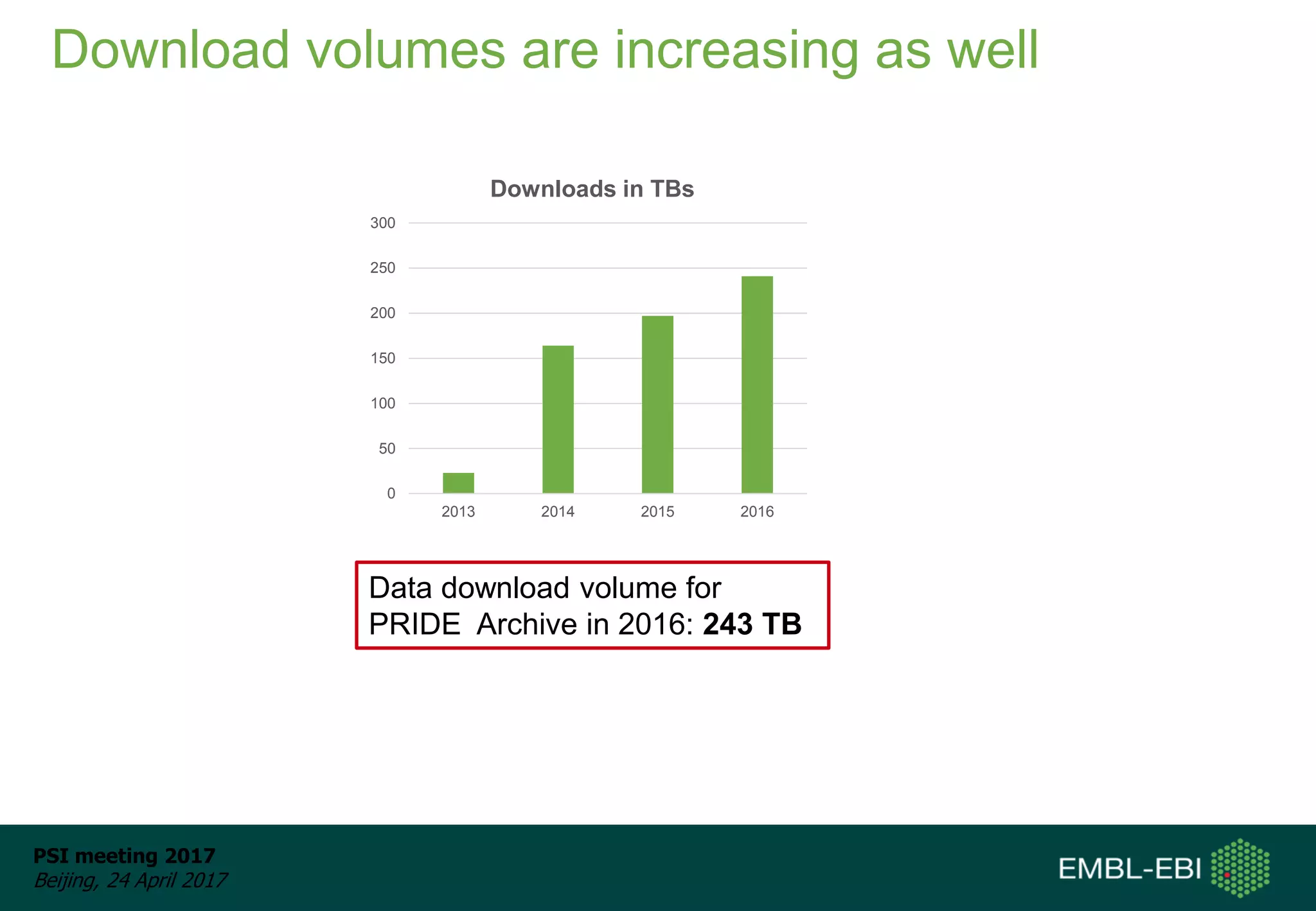
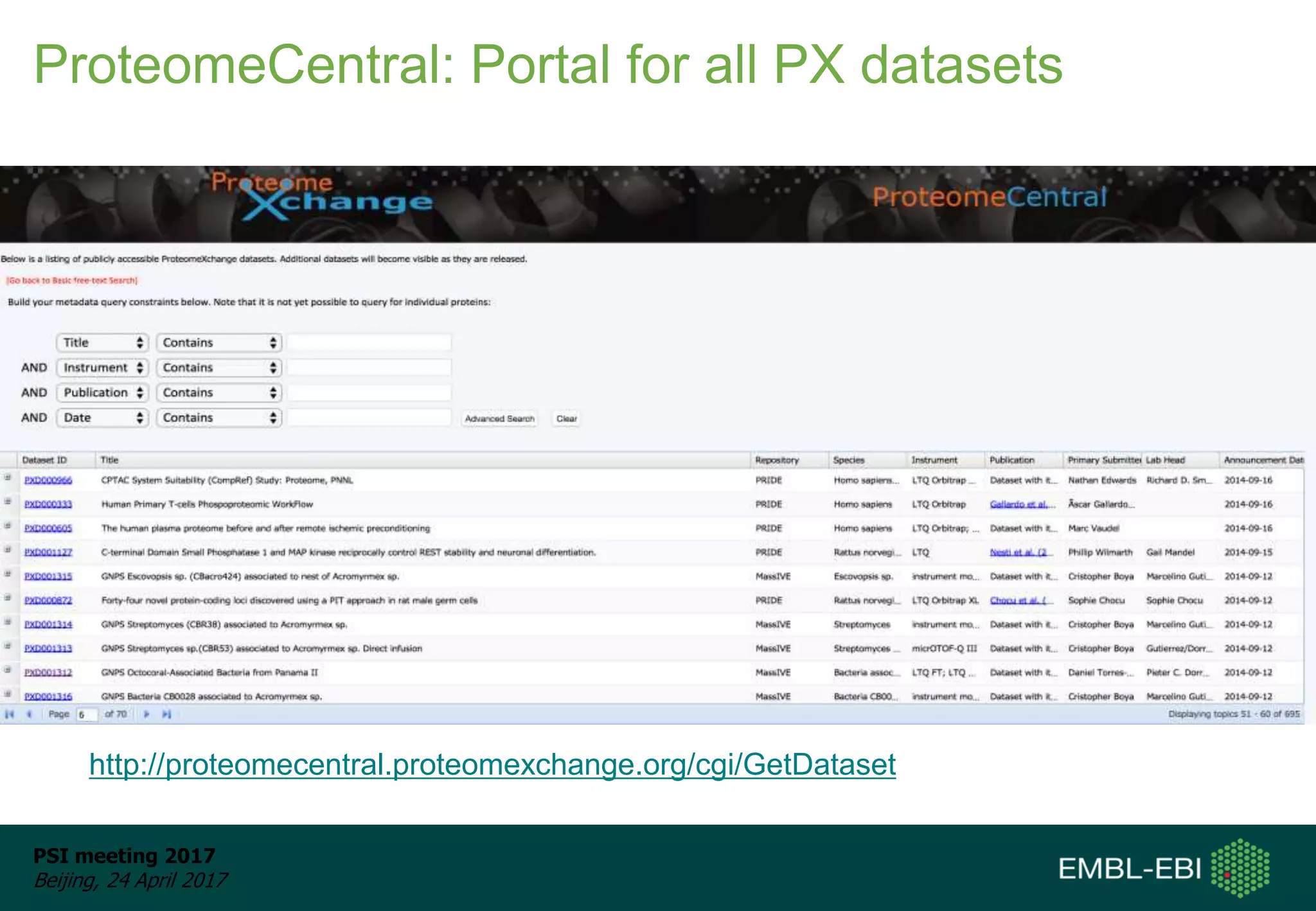
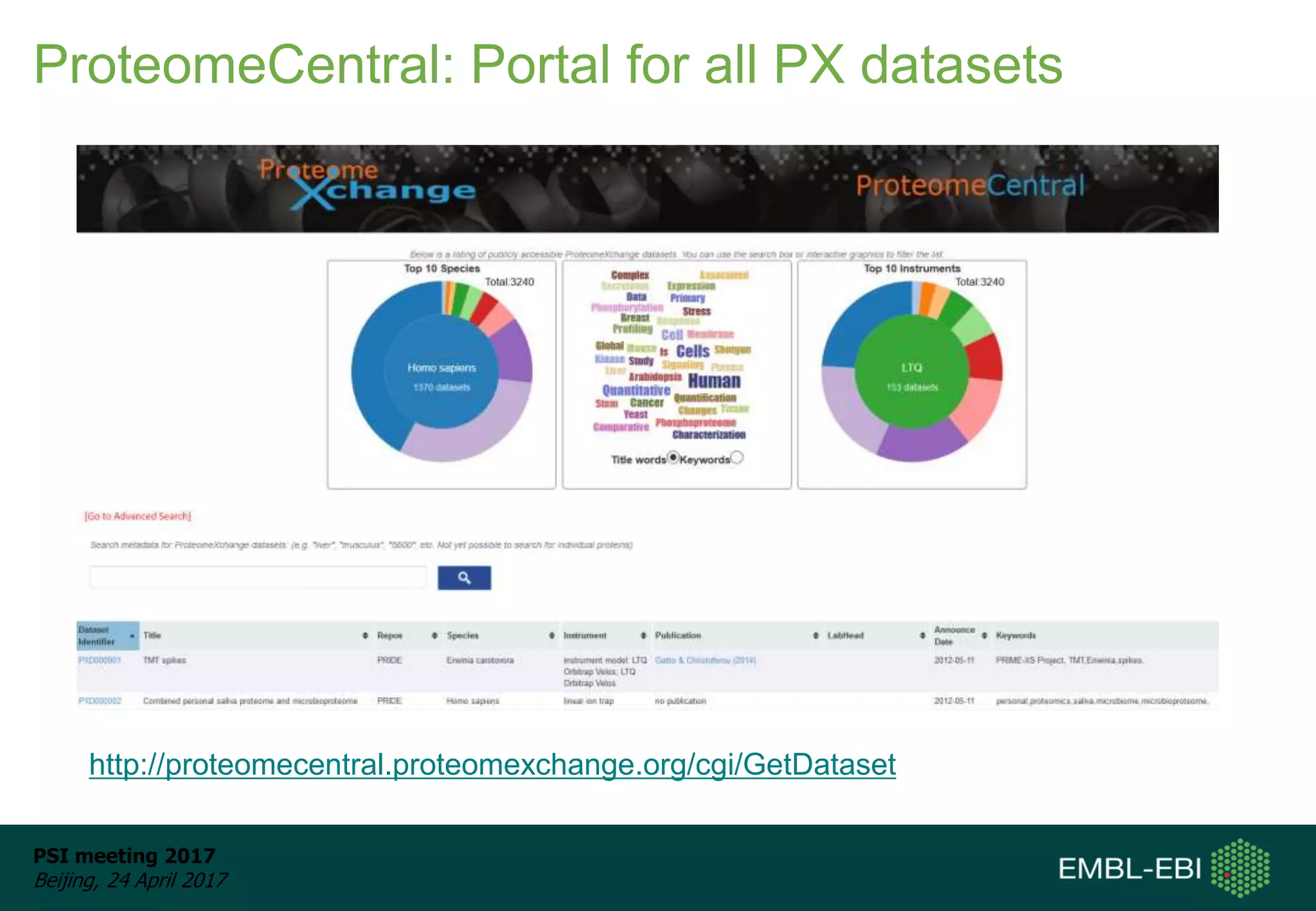
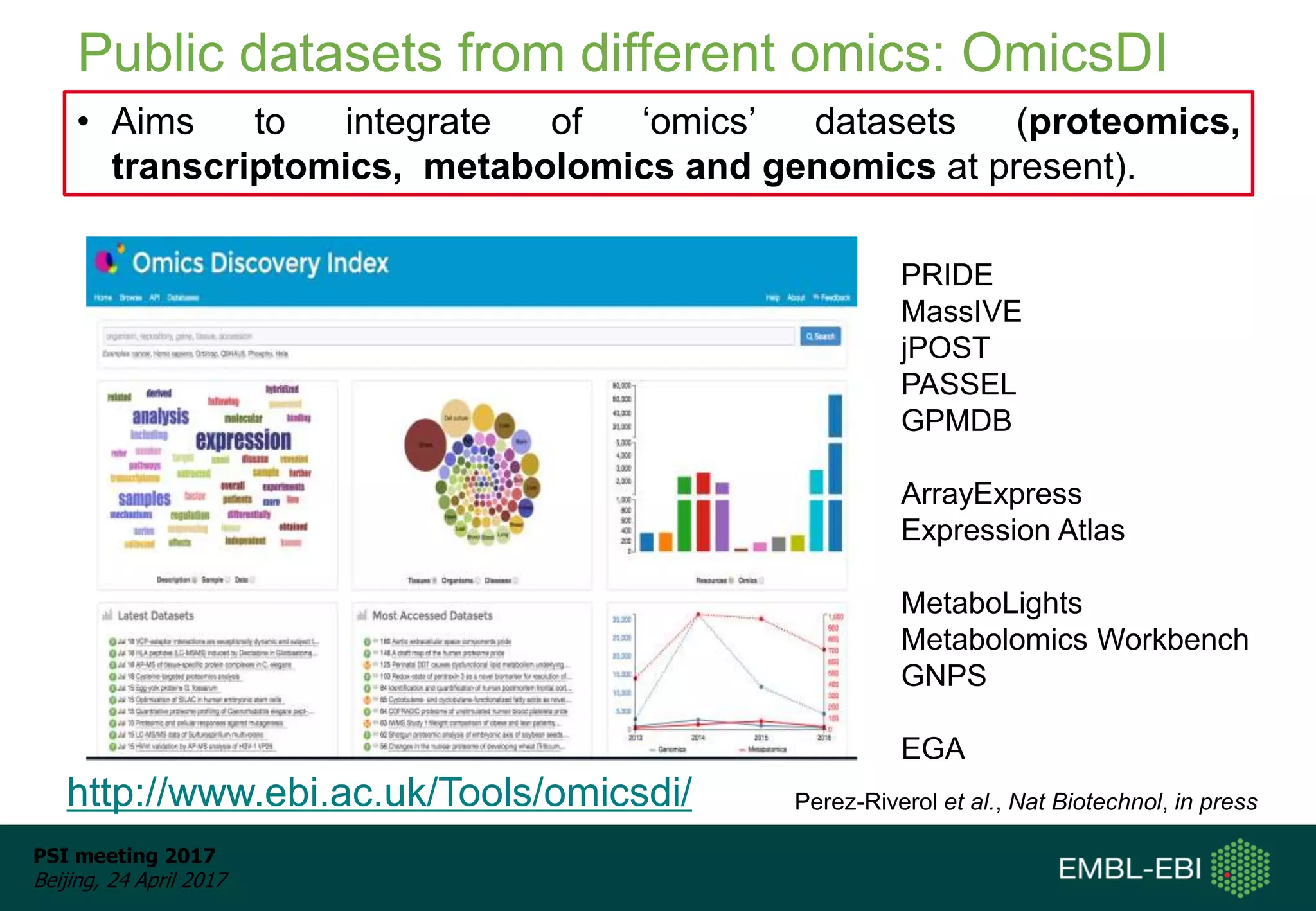
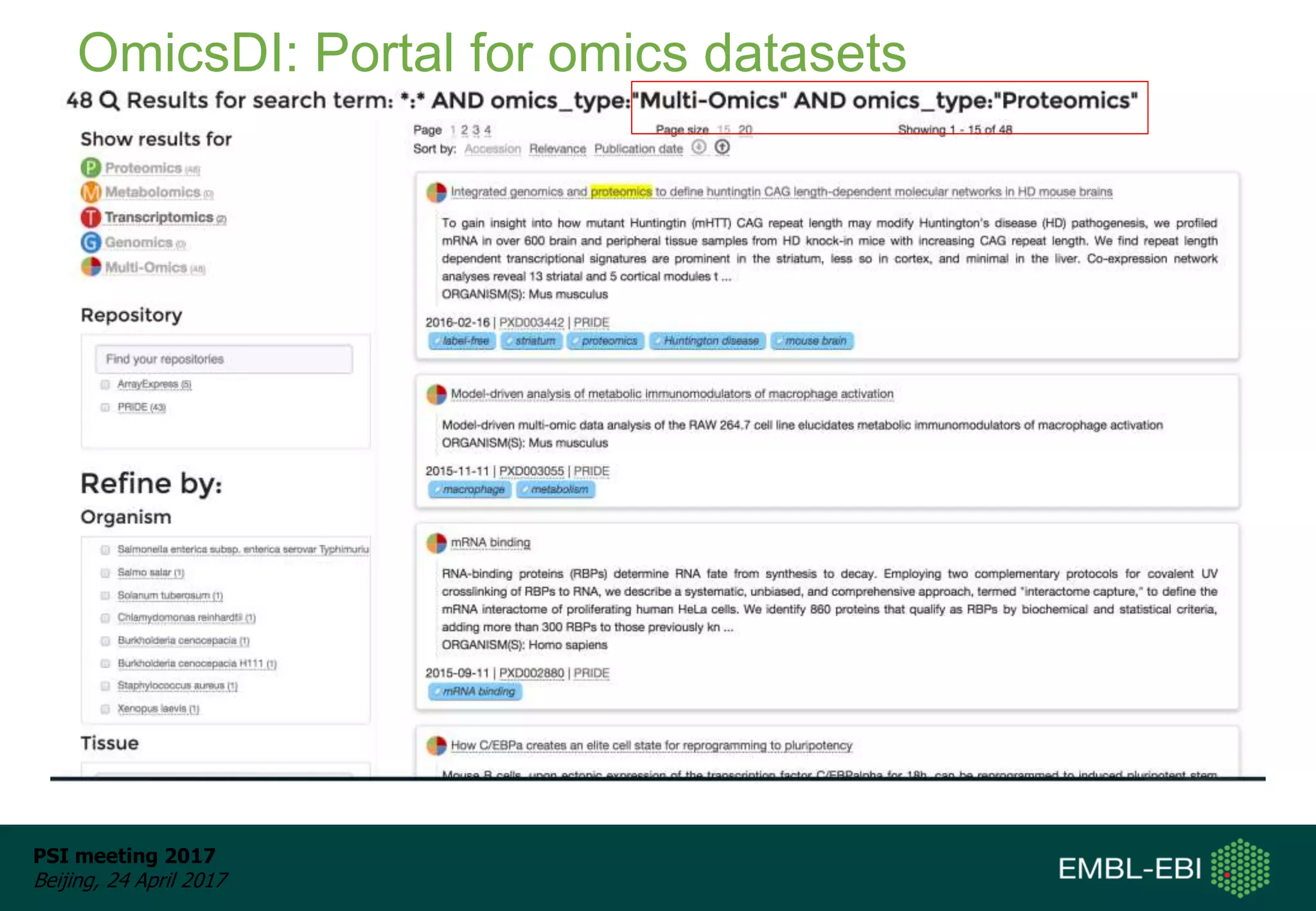
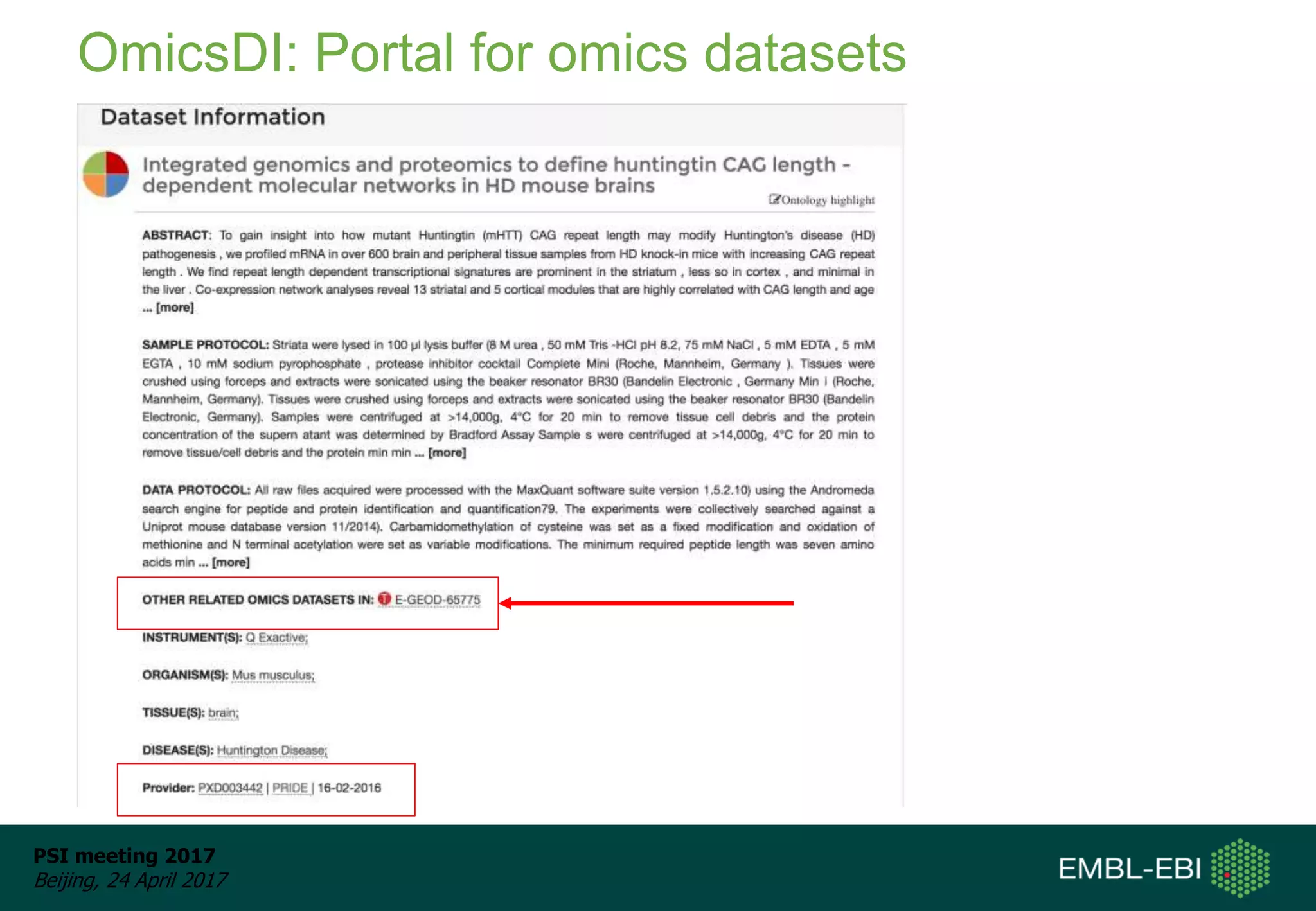
This document provides an overview and status update of ProteomeXchange in 2017. It discusses submission and download statistics showing growth in datasets submitted. There are now over 5,000 datasets in PRIDE from over 1,000 species. Download volumes have increased to over 200 TB in 2016. Citations of proteomics datasets are also increasing. A new prospective member, Firmiana, may join ProteomeXchange. The OmicsDI interface provides integrated access to datasets across multiple omics domains like proteomics, transcriptomics and metabolomics.