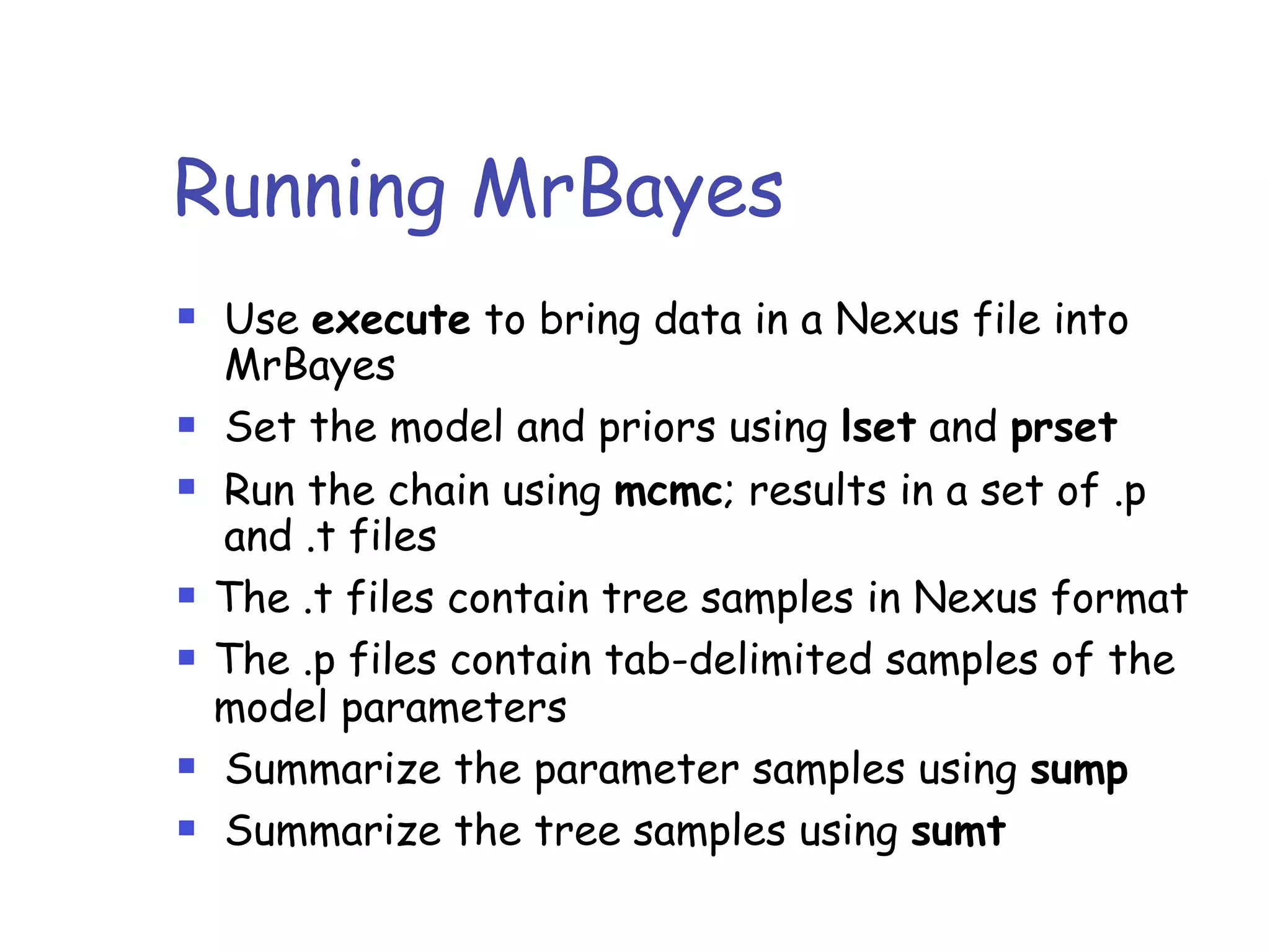
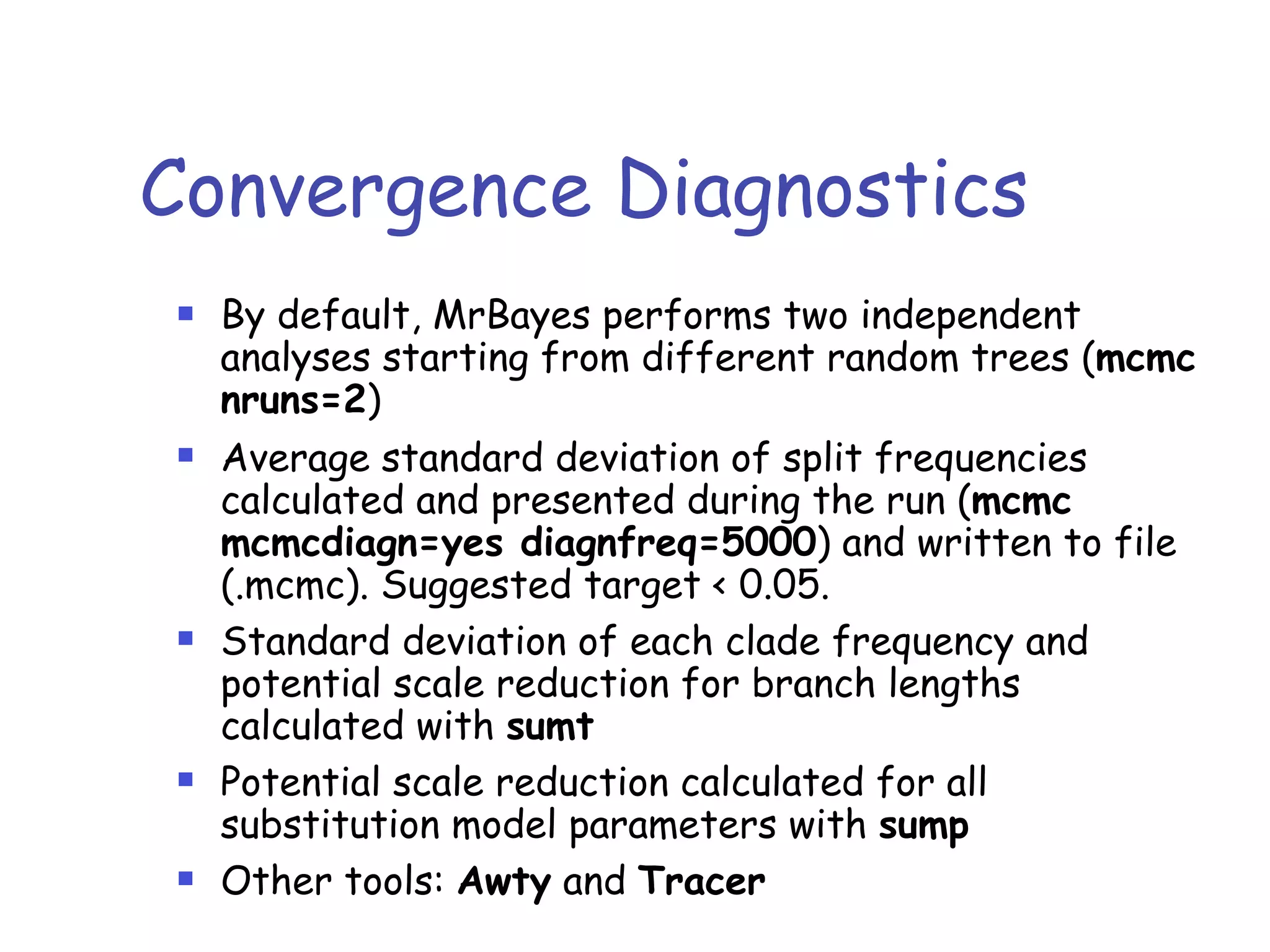
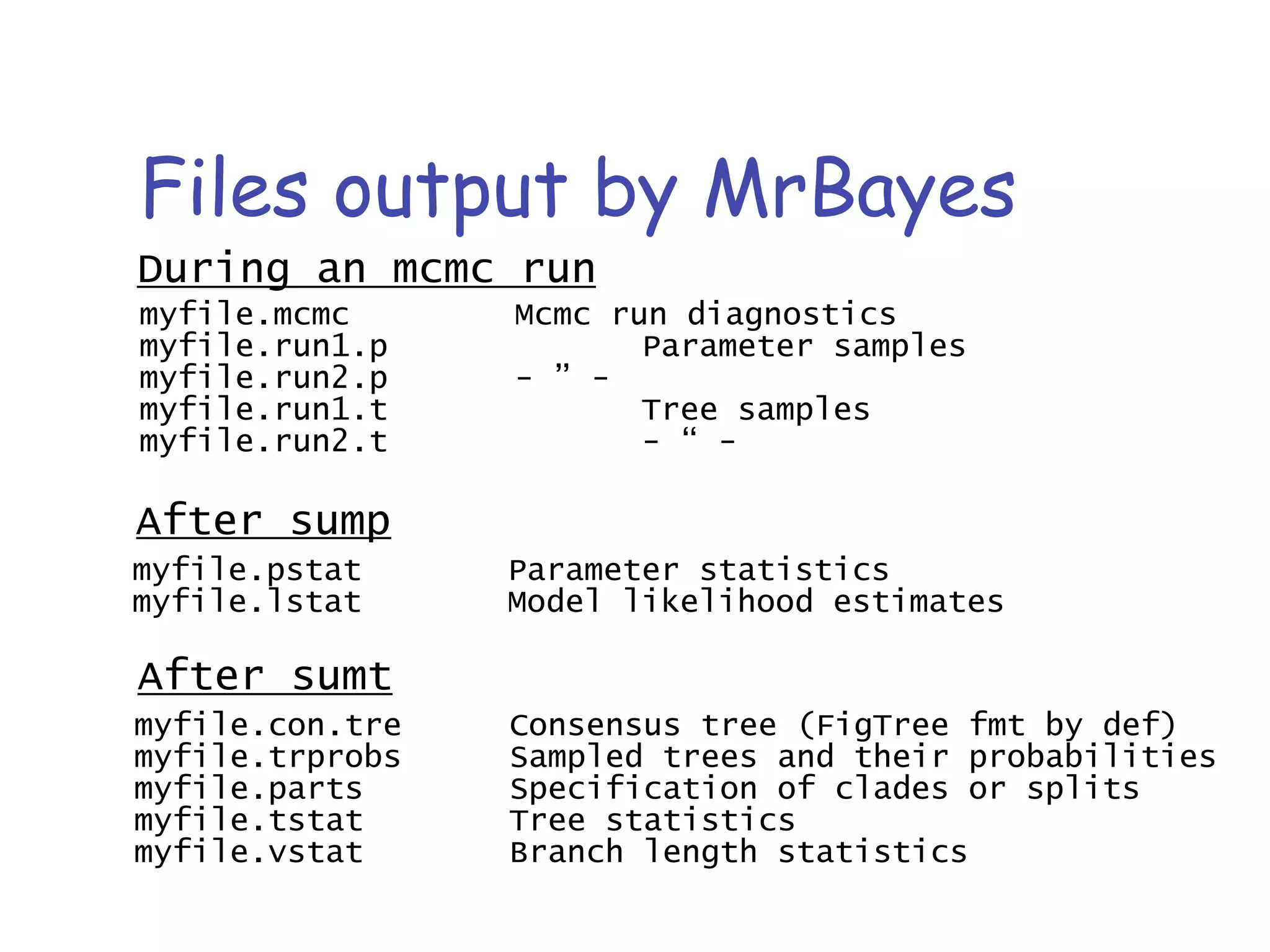
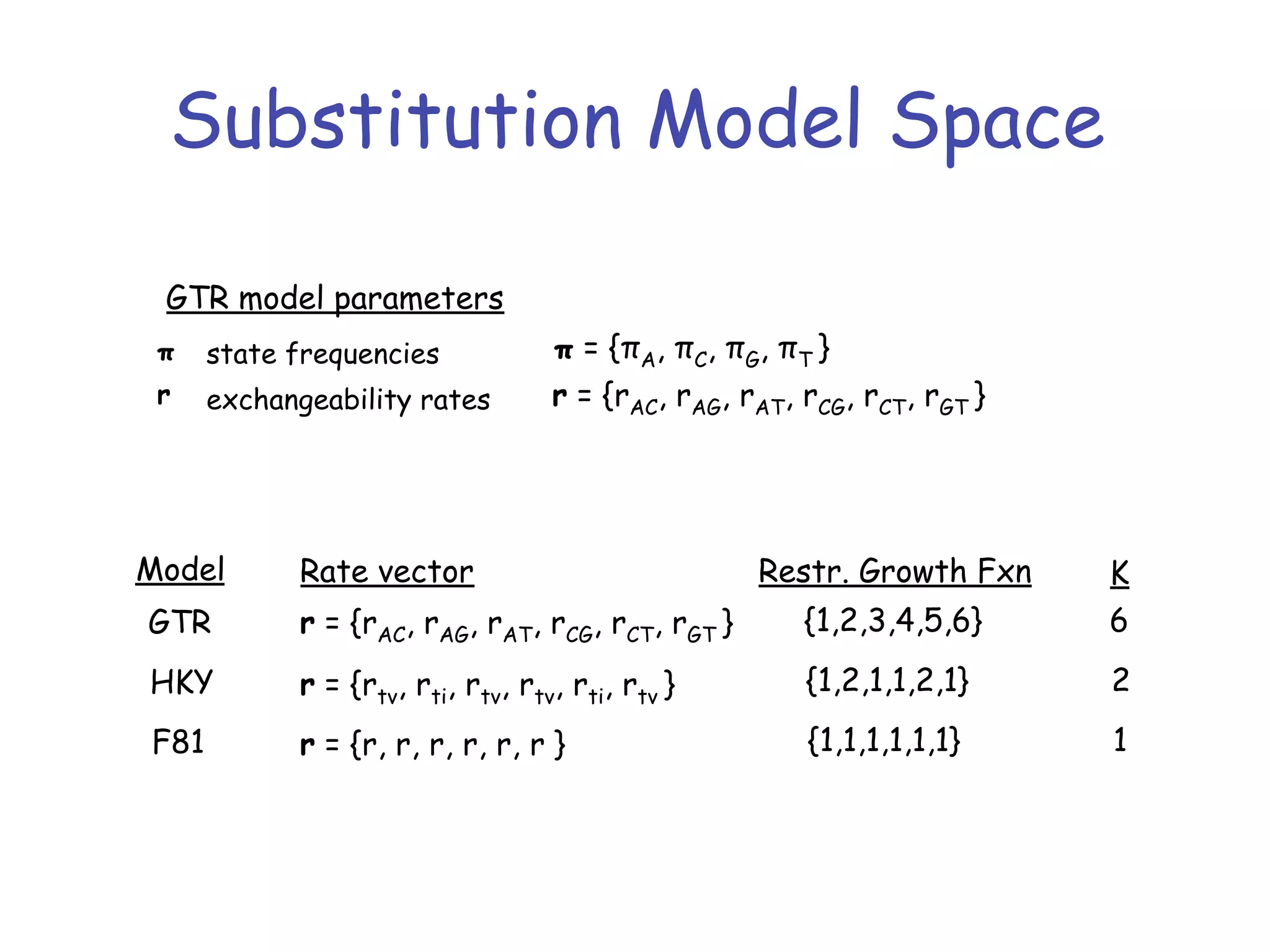
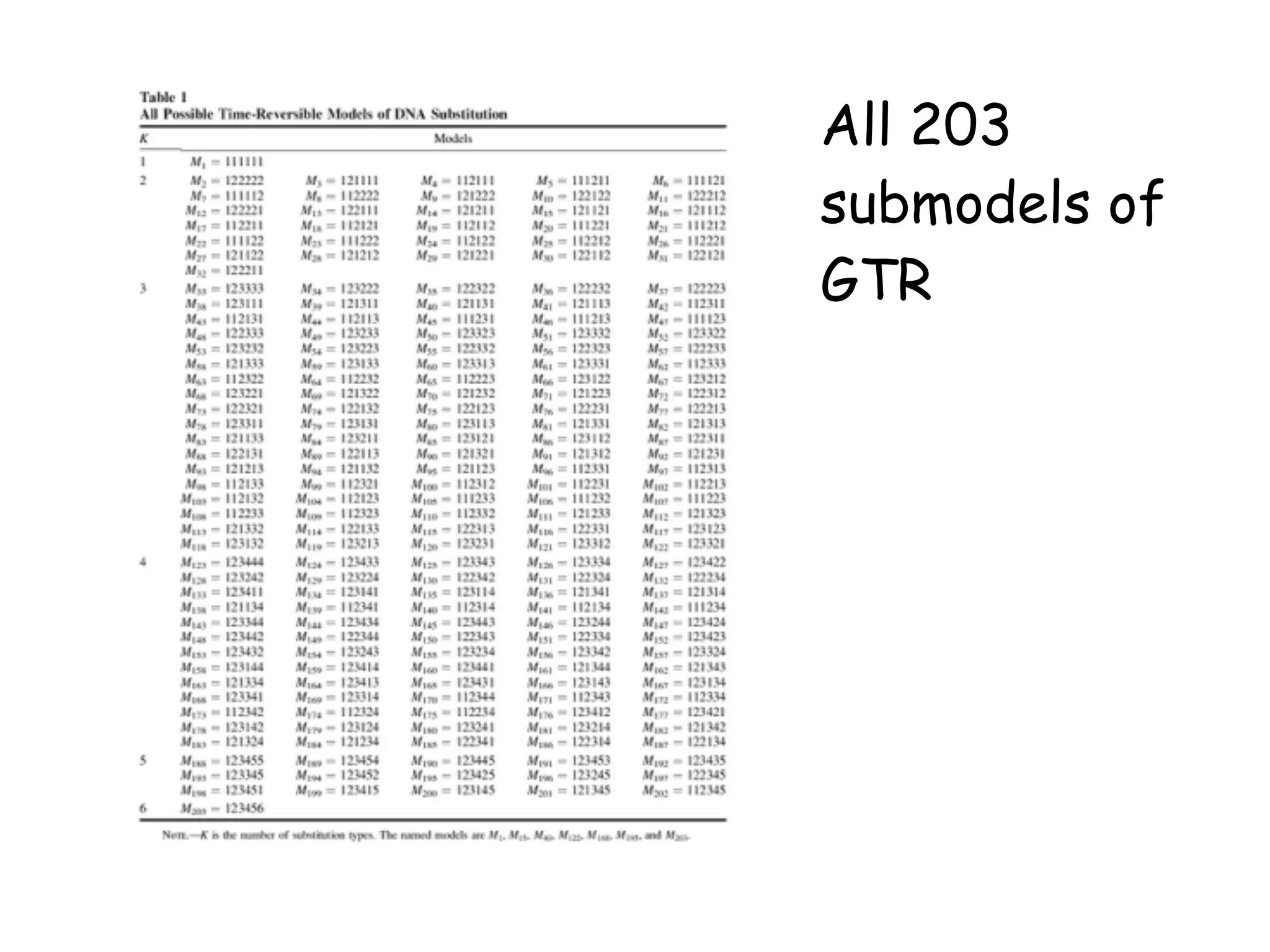
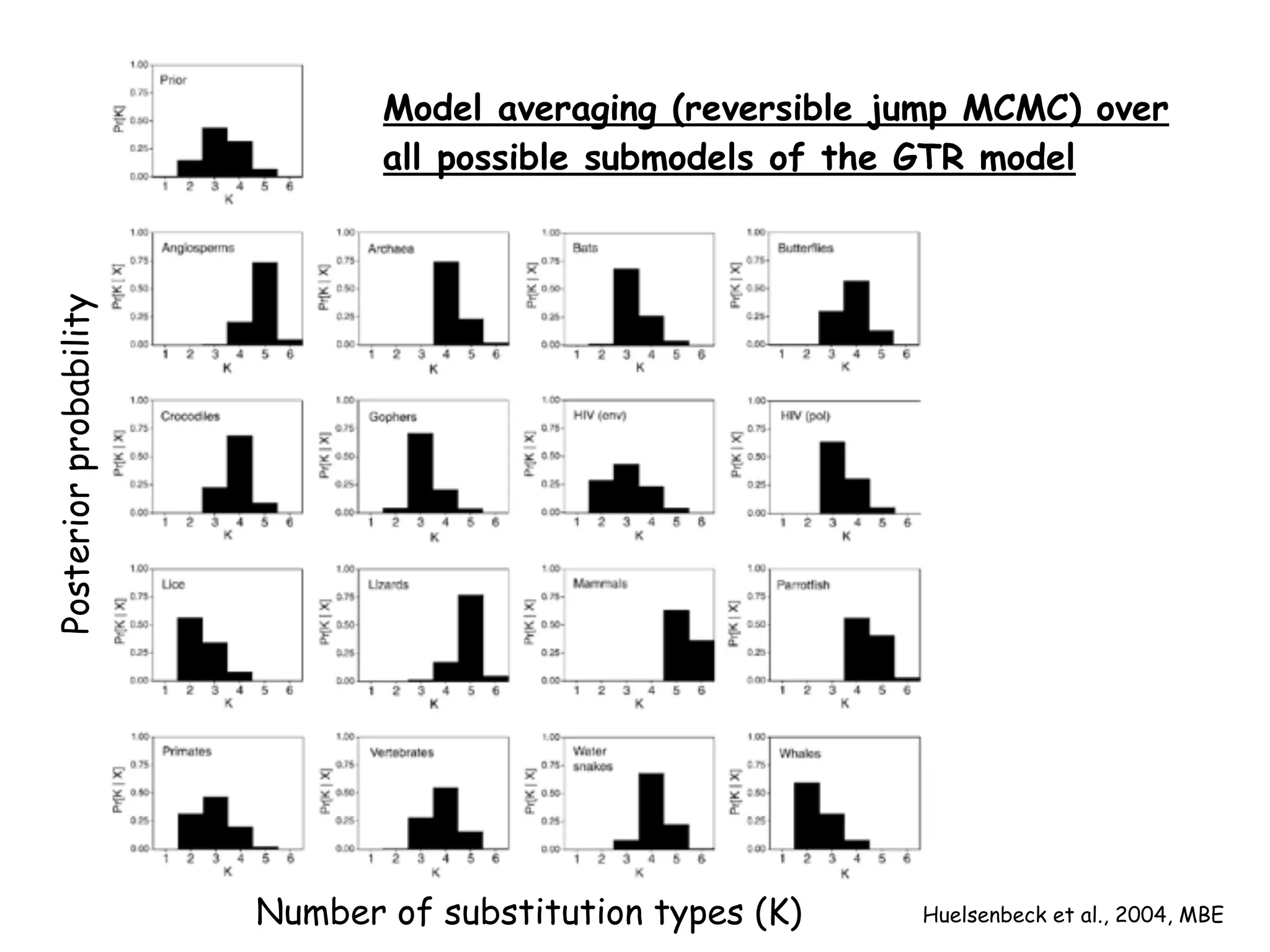
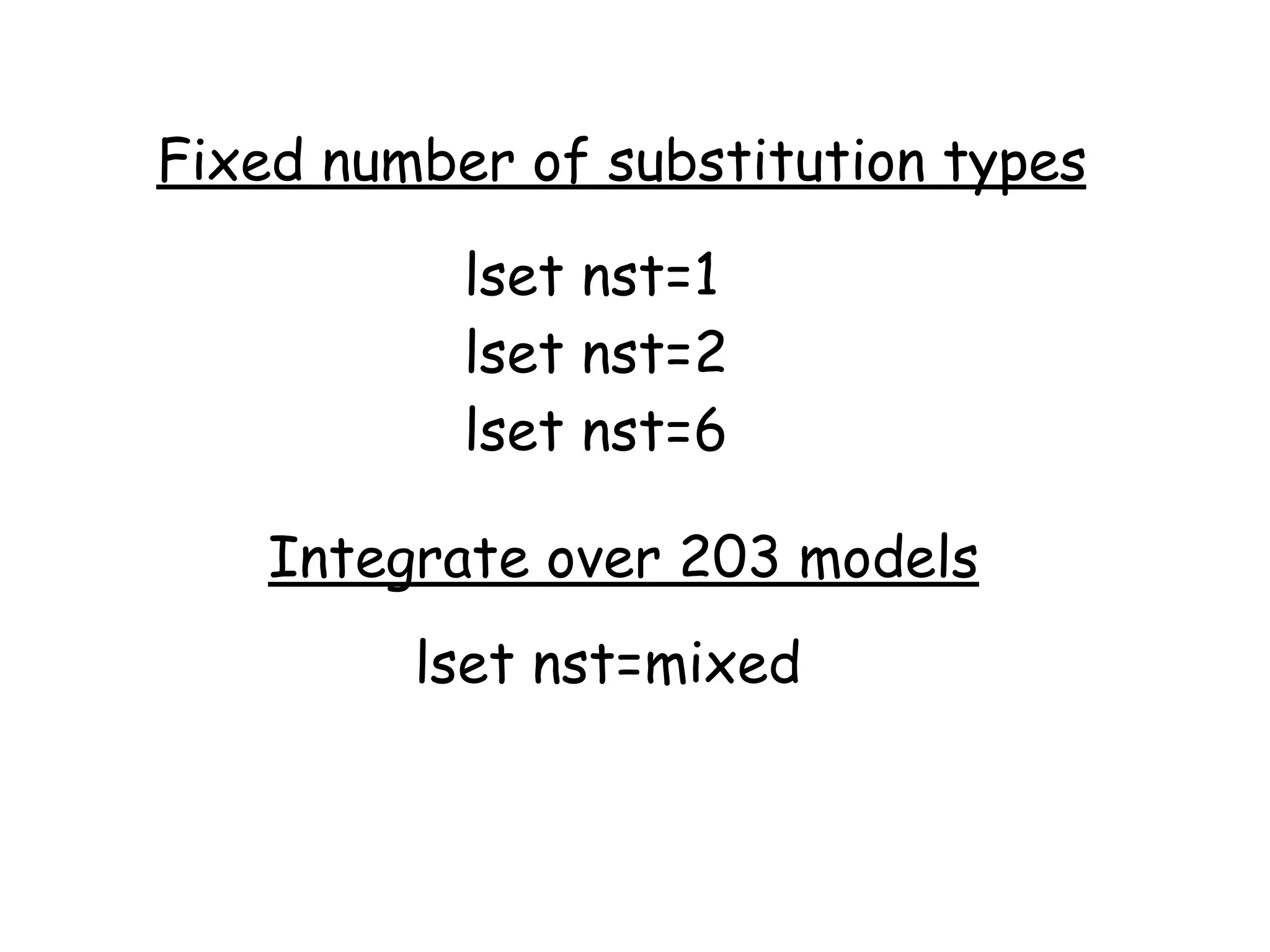
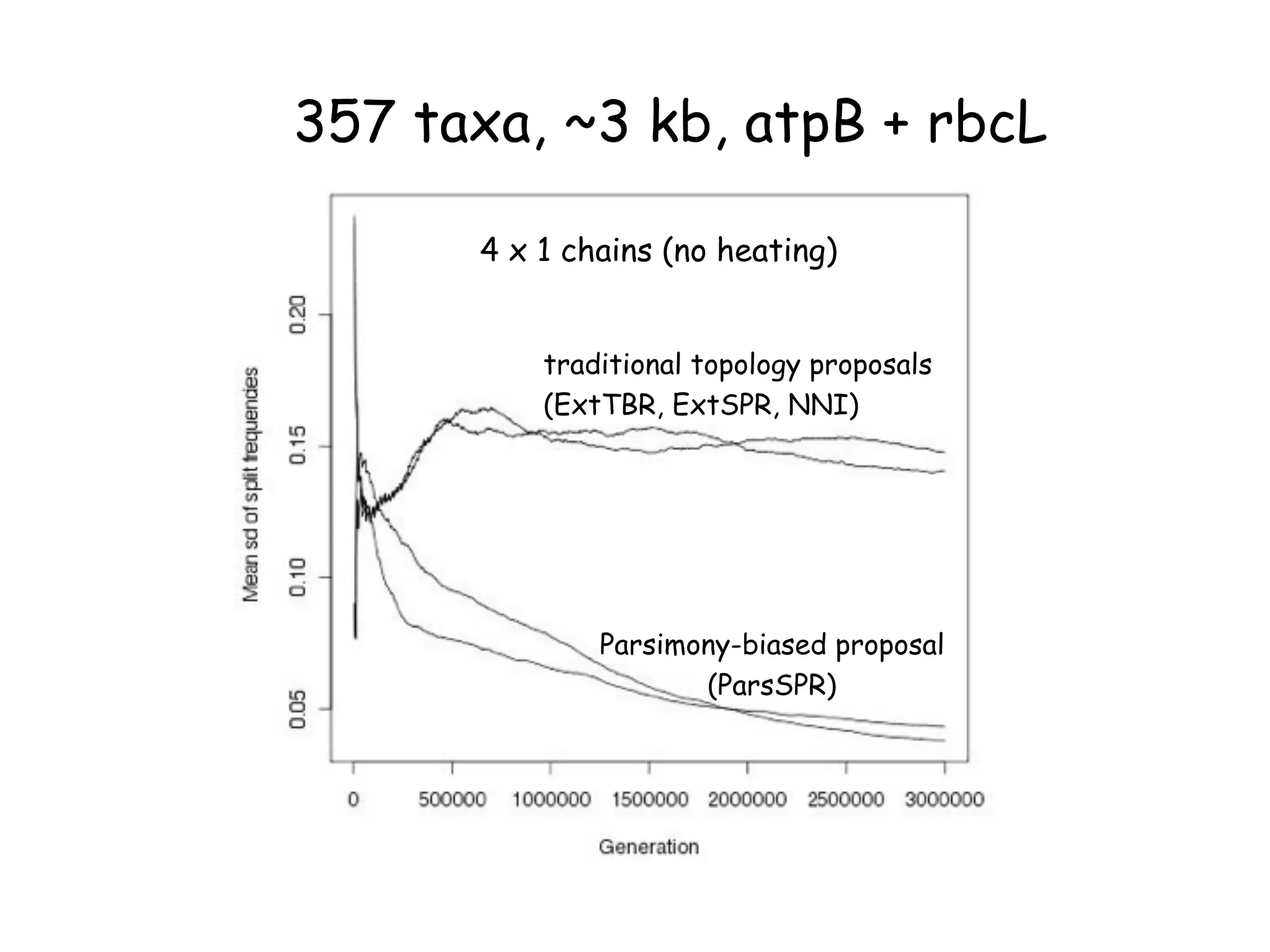
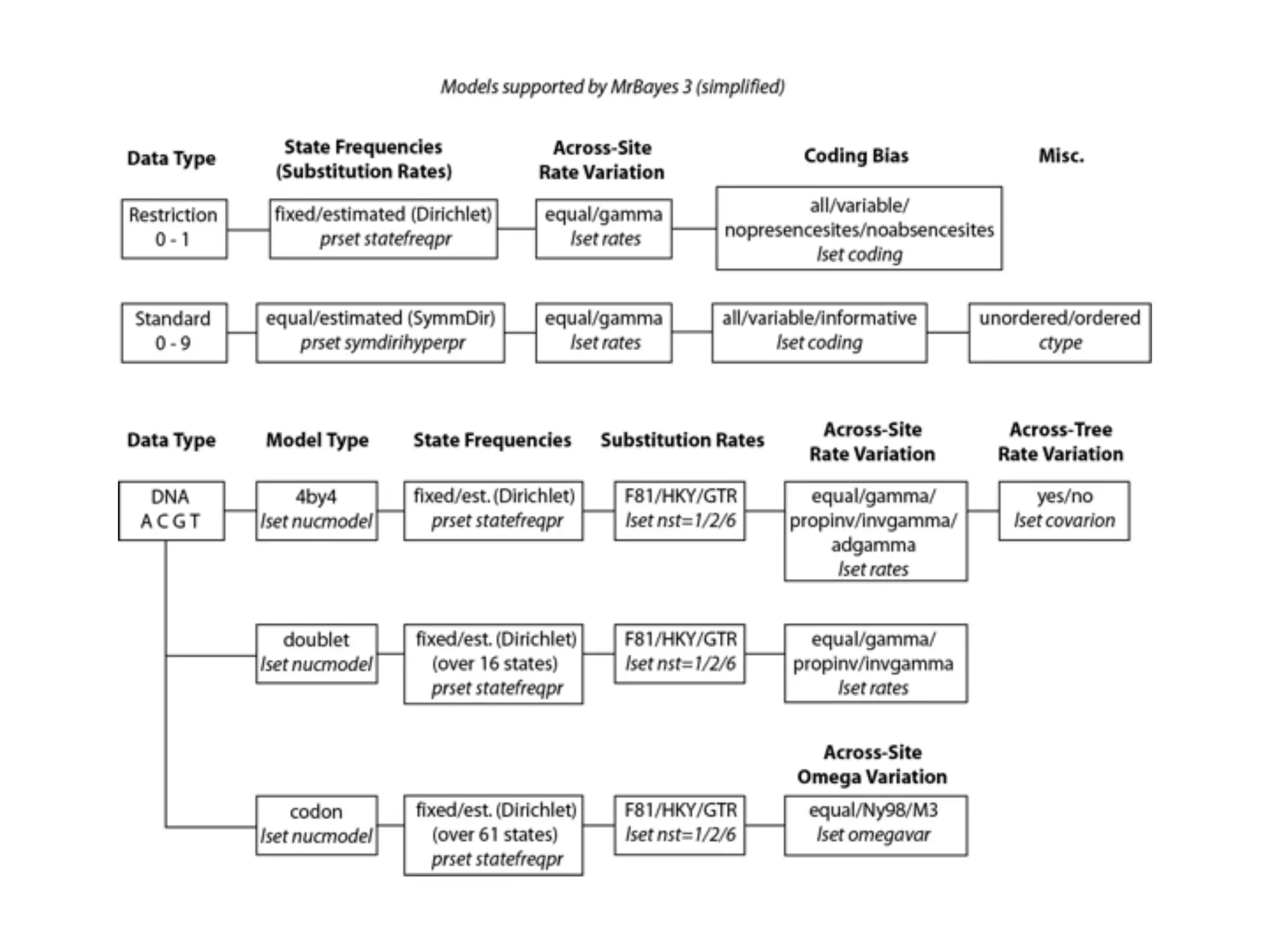
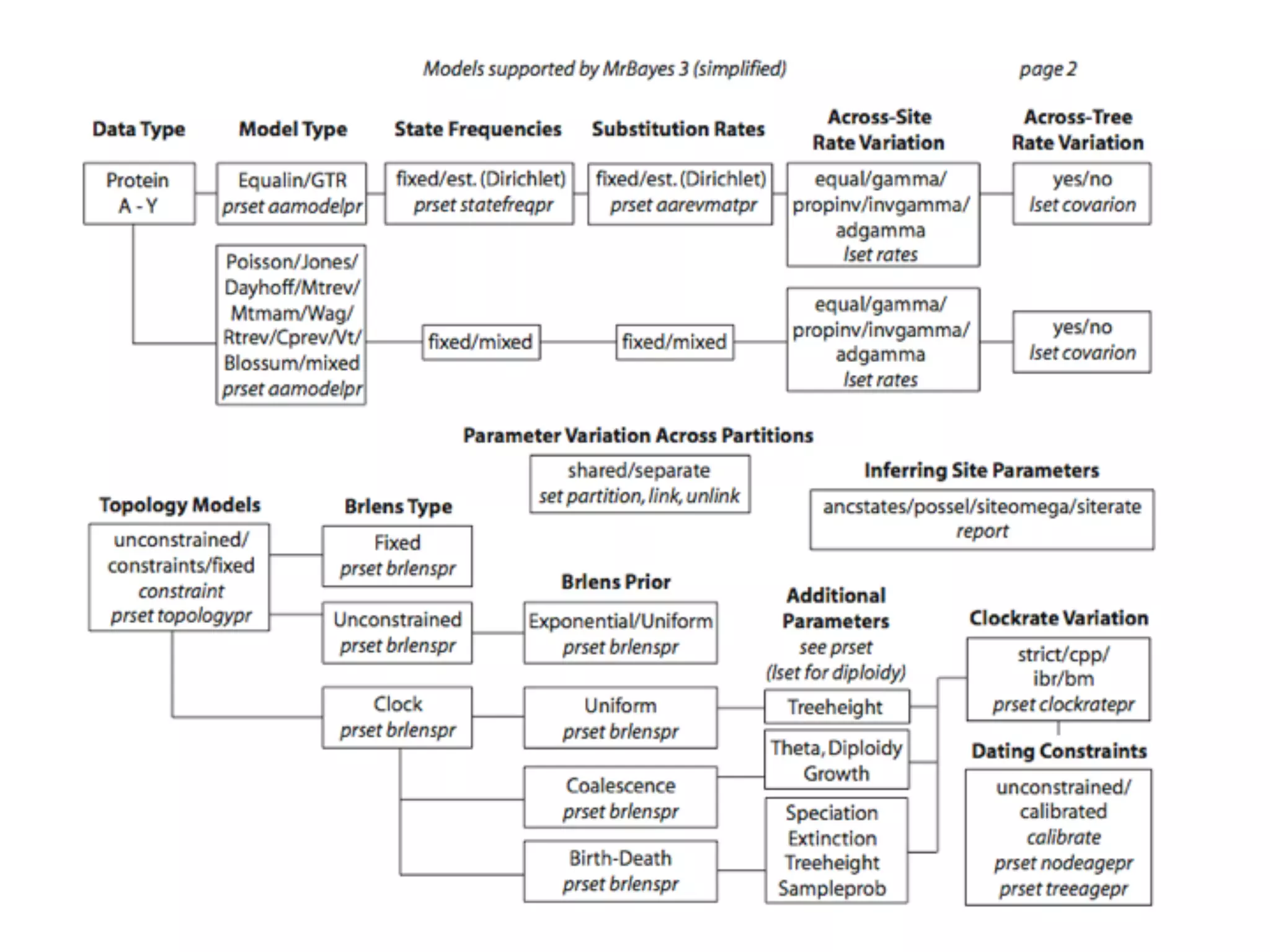
MrBayes is software for Bayesian inference of phylogenetic trees. It can be downloaded from mrbayes.net or compiled from source on GitHub. To run an analysis, users bring data into MrBayes, set the model and priors, run an MCMC chain to sample trees and parameters, and summarize the results. MrBayes outputs diagnostic files during runs and files summarizing trees and parameters after runs complete. Version 3.2 added features like topological constraints, relaxed clocks, and model averaging across substitution models. The manual provides tutorials and guidance on performing analyses and interpreting results.