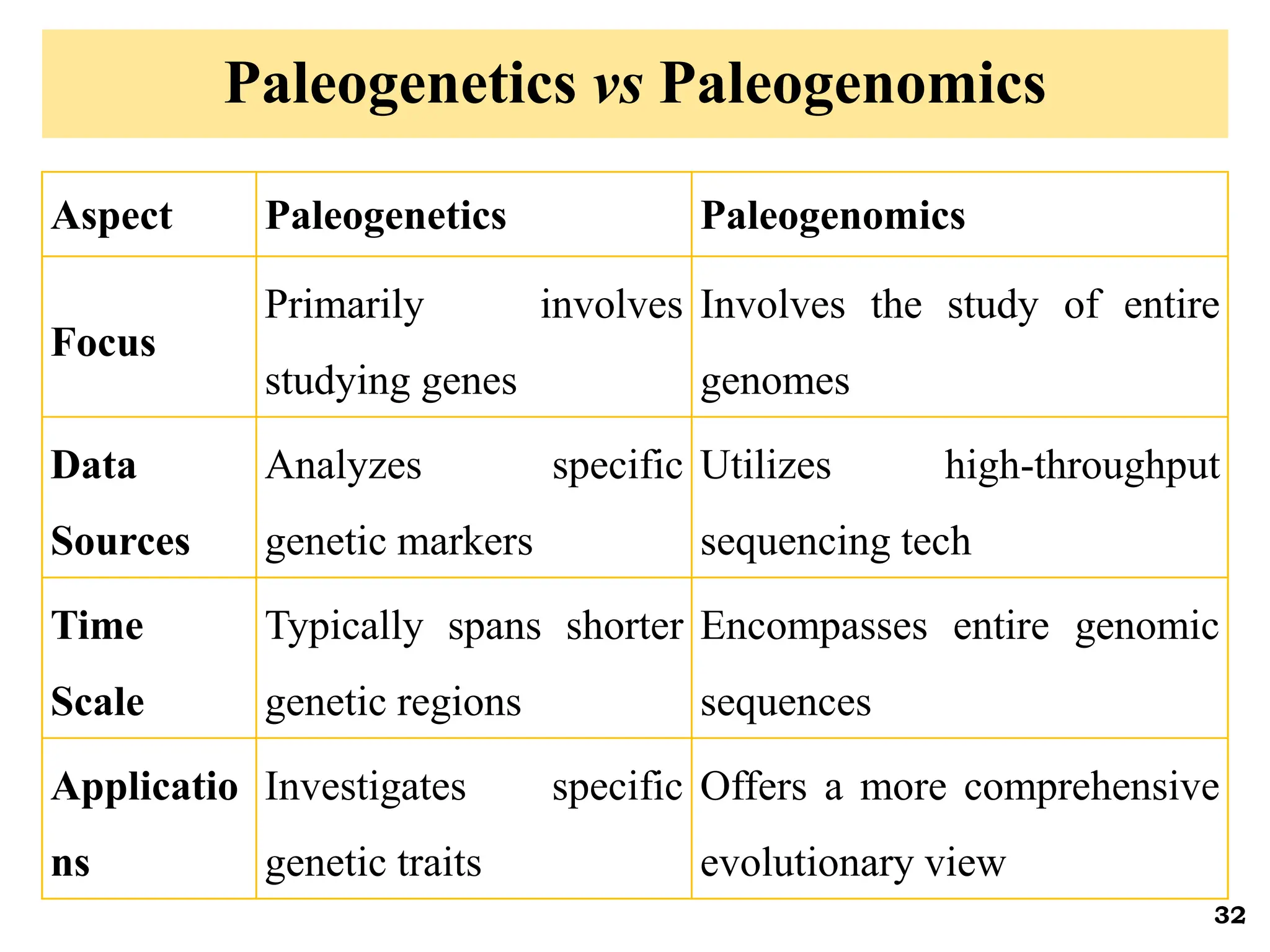
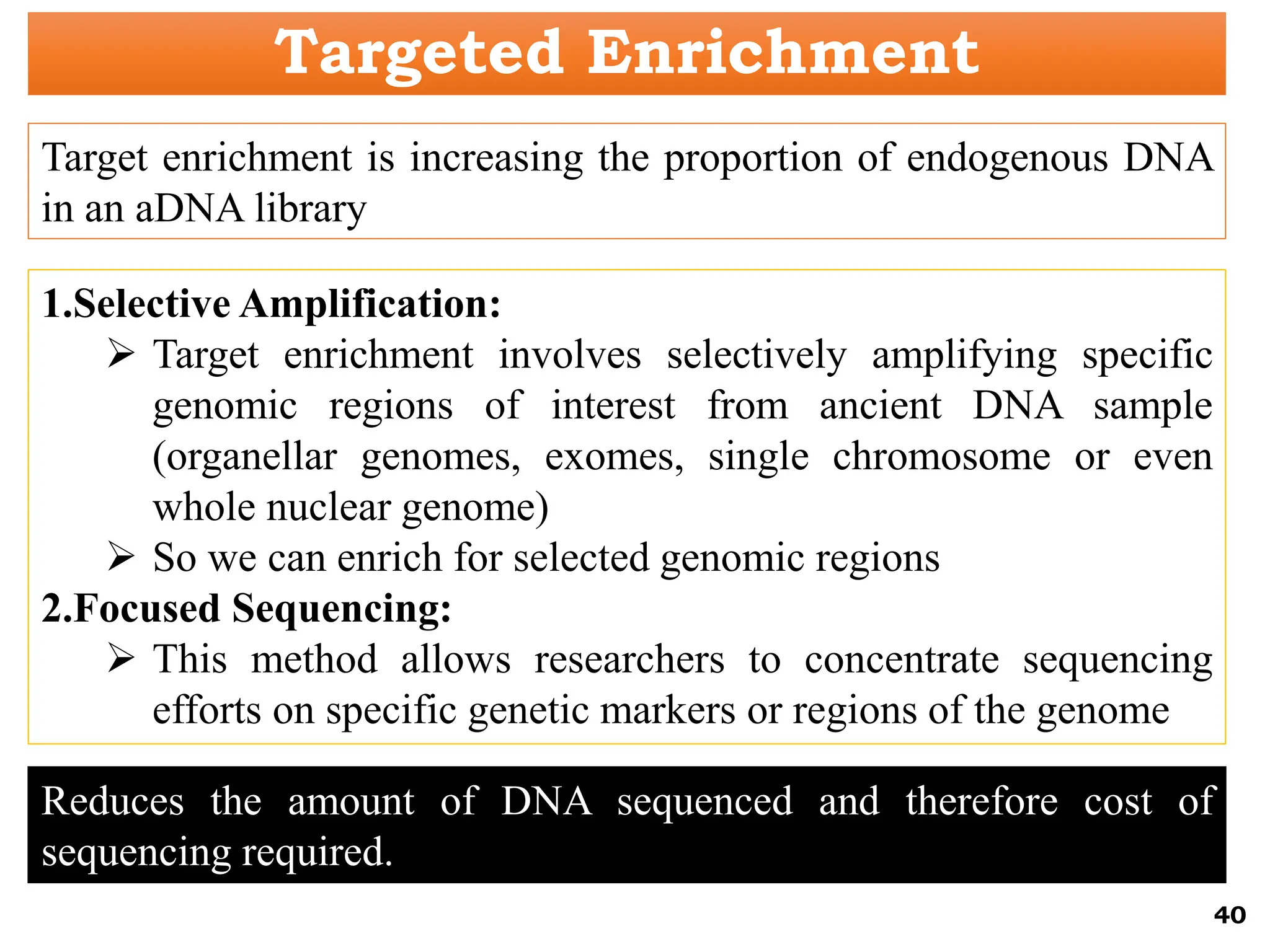
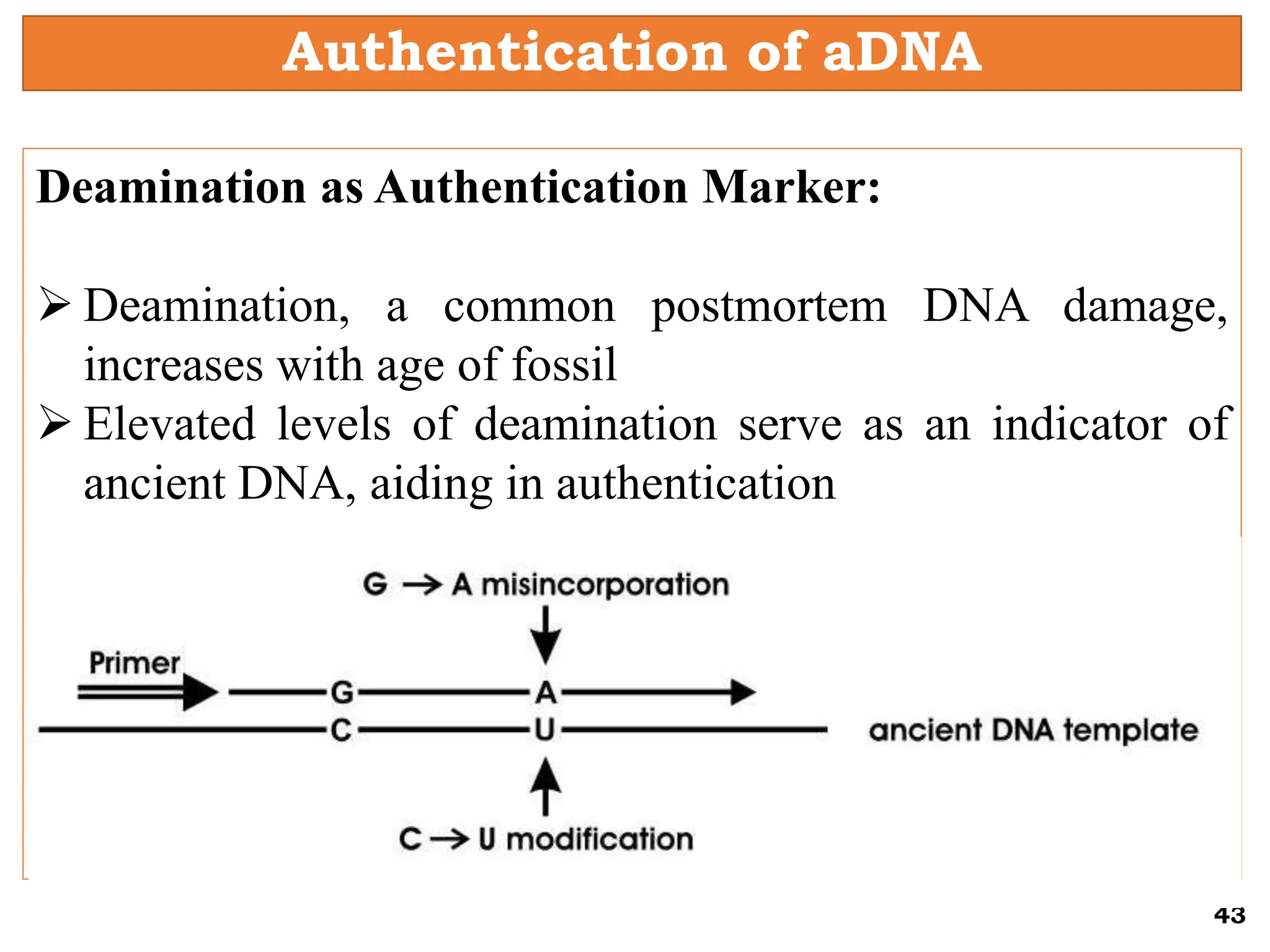
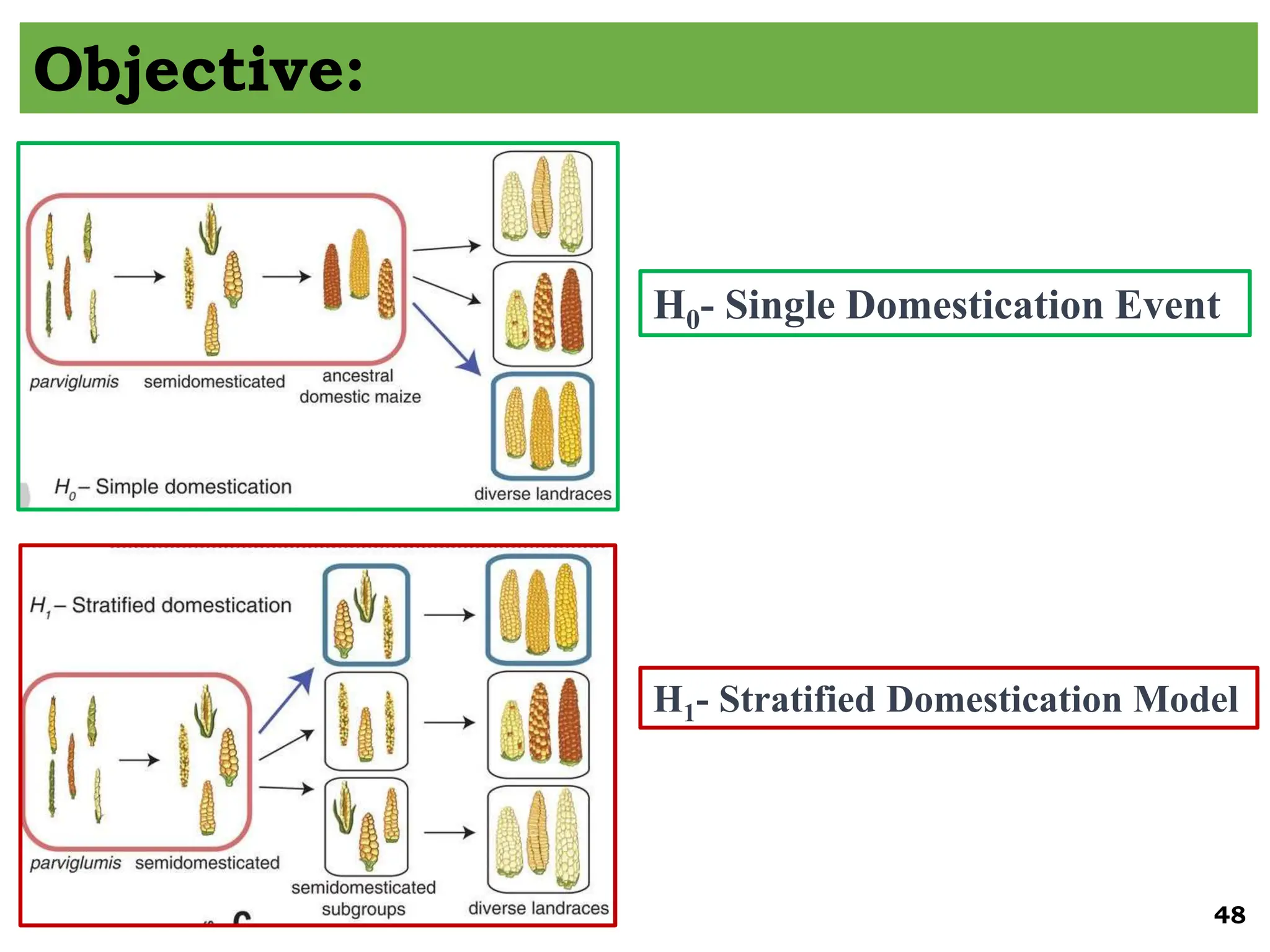
The document discusses the study of paleogenomics, focusing on the evolutionary history of plant species through fossil records, genetic analysis, and living organisms' genetic structures. It highlights the challenges of analyzing ancient DNA and elaborates on methods of extracting and sequencing this DNA to understand ancient ecosystems and the evolution of various species. Key case studies, such as maize domestication, illustrate the complexities involved in plant evolution and adaptation over time.

















![Ancient Genome Reconstruction using Synchronic
Approach
Ortholog
identification
→ pPGs
[Putative protogenes]
Synteny identification
→SBs
[Synteny blocks]
Ancestral chromosome
→ Core-pPGs
[Core protogenes]
Ancestral genome
→ oPGs
[Ordered protogenes]
(Pont et al., 2019) 19](https://image.slidesharecdn.com/pamb1066paleogenomics-240801032404-bf0e1163/75/Paleogenomics-Connecting-the-dots-of-crop-evolution-19-2048.jpg)