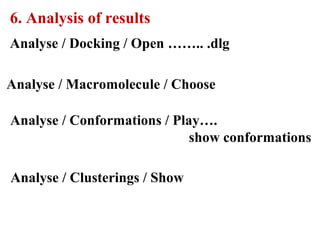
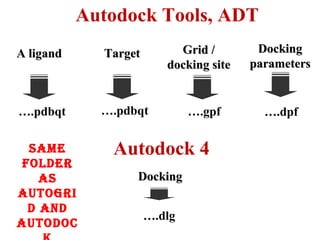
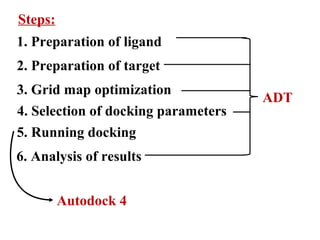
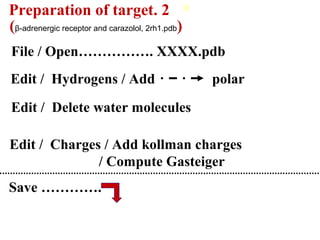
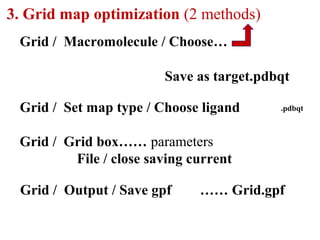
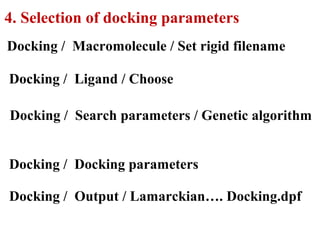
Molecular docking software Autodock 4.2.6 is presented. It is free and credible for predicting optimal bound conformations of ligands to proteins. The steps for using Autodock include: (1) preparing the ligand and target protein, (2) optimizing a grid map, (3) selecting docking parameters, (4) running the docking simulation, and (5) analyzing the results. Autodock Tools (ADT) is used for preparation of files in .pdbqt format before docking with Autodock 4.




![5. Running Docking
Start menu / Run ….. cmd.exe
Command lines:
cd folder name/ path
autogrid4 –p grid.gpf [-l grid.glg]
autodock4 –p dock.dpf [-l dock.dlg]](https://image.slidesharecdn.com/autodock-slideshare-170501121446/85/Molecular-Docking-using-Autodock-4-2-6-10-320.jpg)