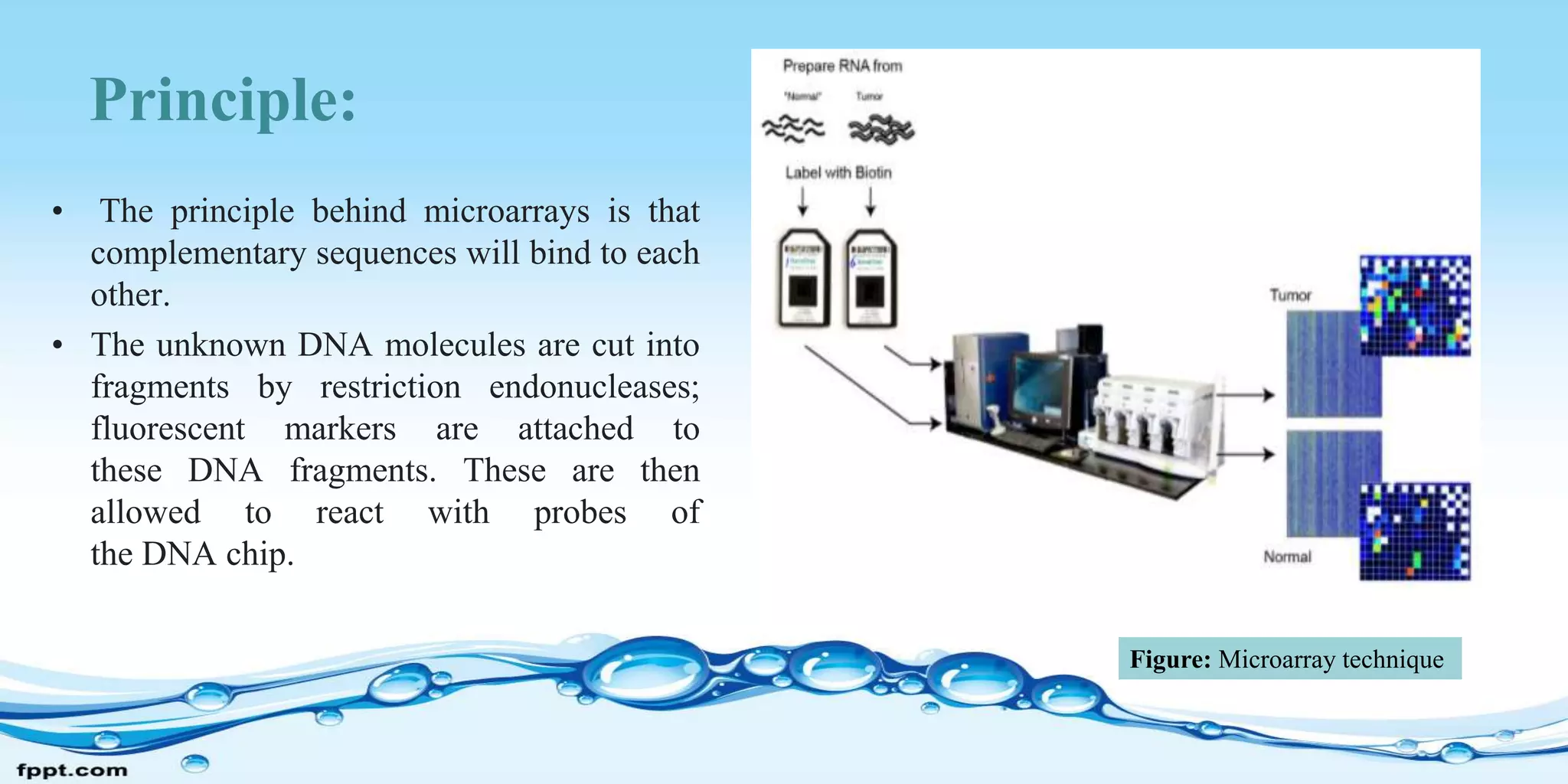
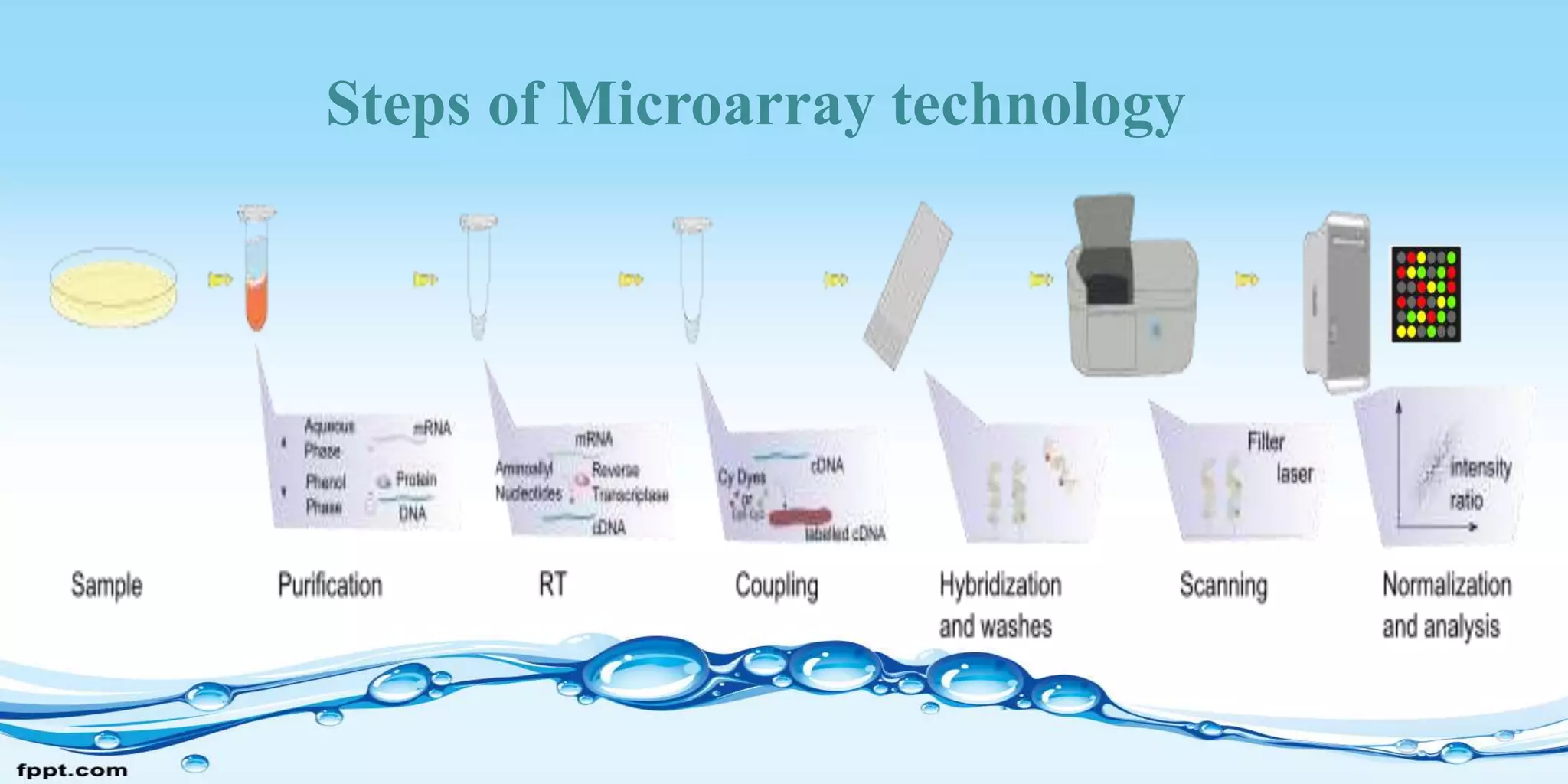
Microarray technology allows researchers to study the expression of thousands of genes simultaneously. It involves placing short DNA sequences from known genes onto a glass slide in a controlled array. Researchers apply fluorescent-tagged DNA or RNA samples to the array to see which sequences bind, indicating gene expression. Microarrays can be used for gene expression profiling, comparative genomics, disease diagnosis, drug discovery, and toxicology research. They allow analysis of entire genomes quickly and efficiently. However, microarrays are costly to produce and analyze, and the chips have a limited shelf life.