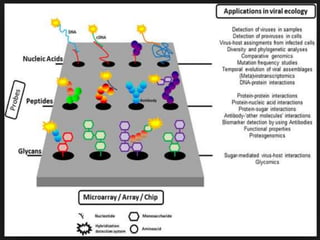
Microarrays can be used for gene expression profiling, comparative genomics, disease diagnosis, drug discovery, and toxicological research. It allows researchers to examine thousands of genes simultaneously and see changes in gene expression patterns. Microarrays have applications in areas like cancer classification, pharmacogenomics, and toxicogenomics. While a powerful tool, microarrays also have limitations like being expensive to create and requiring time to develop.