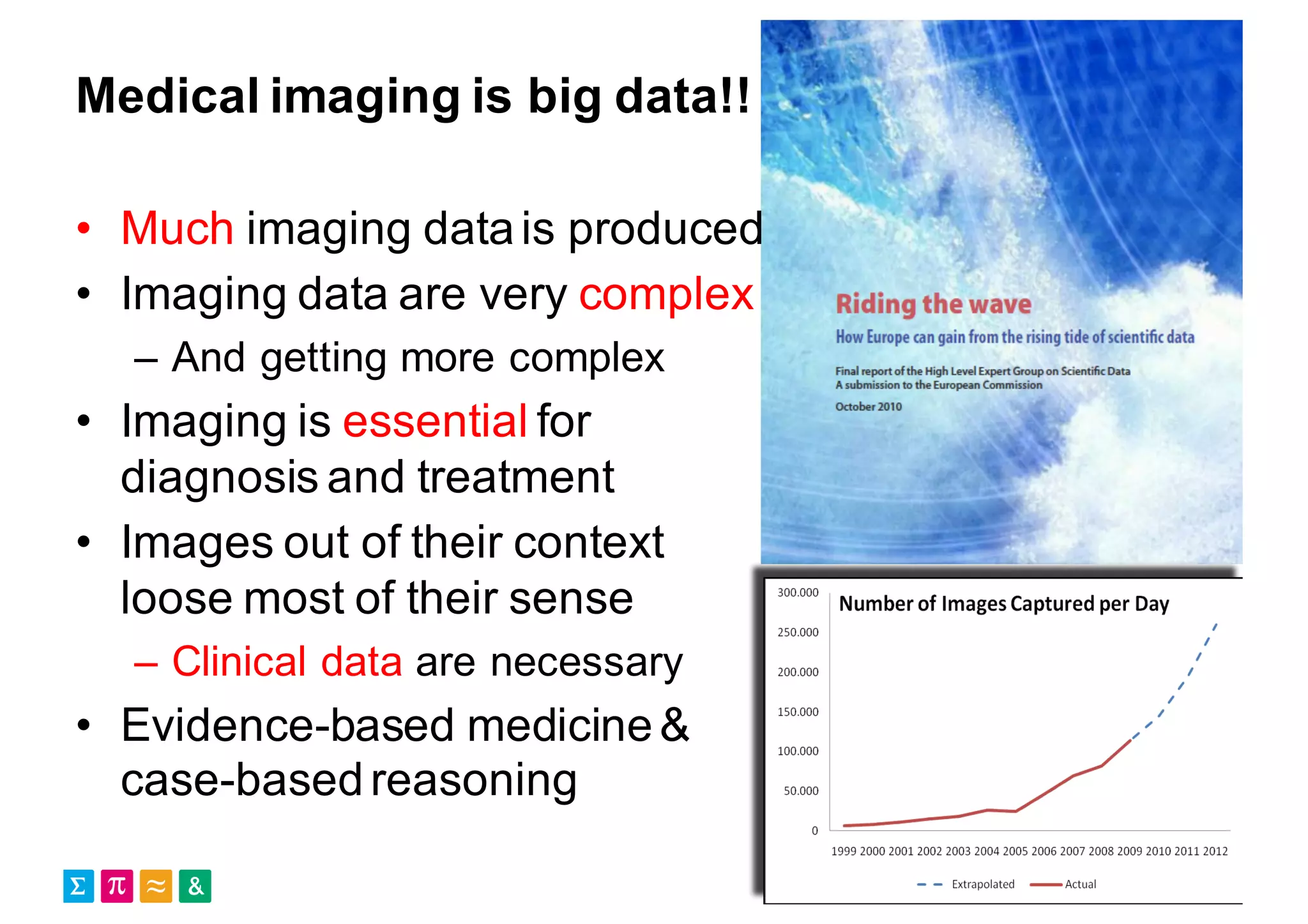
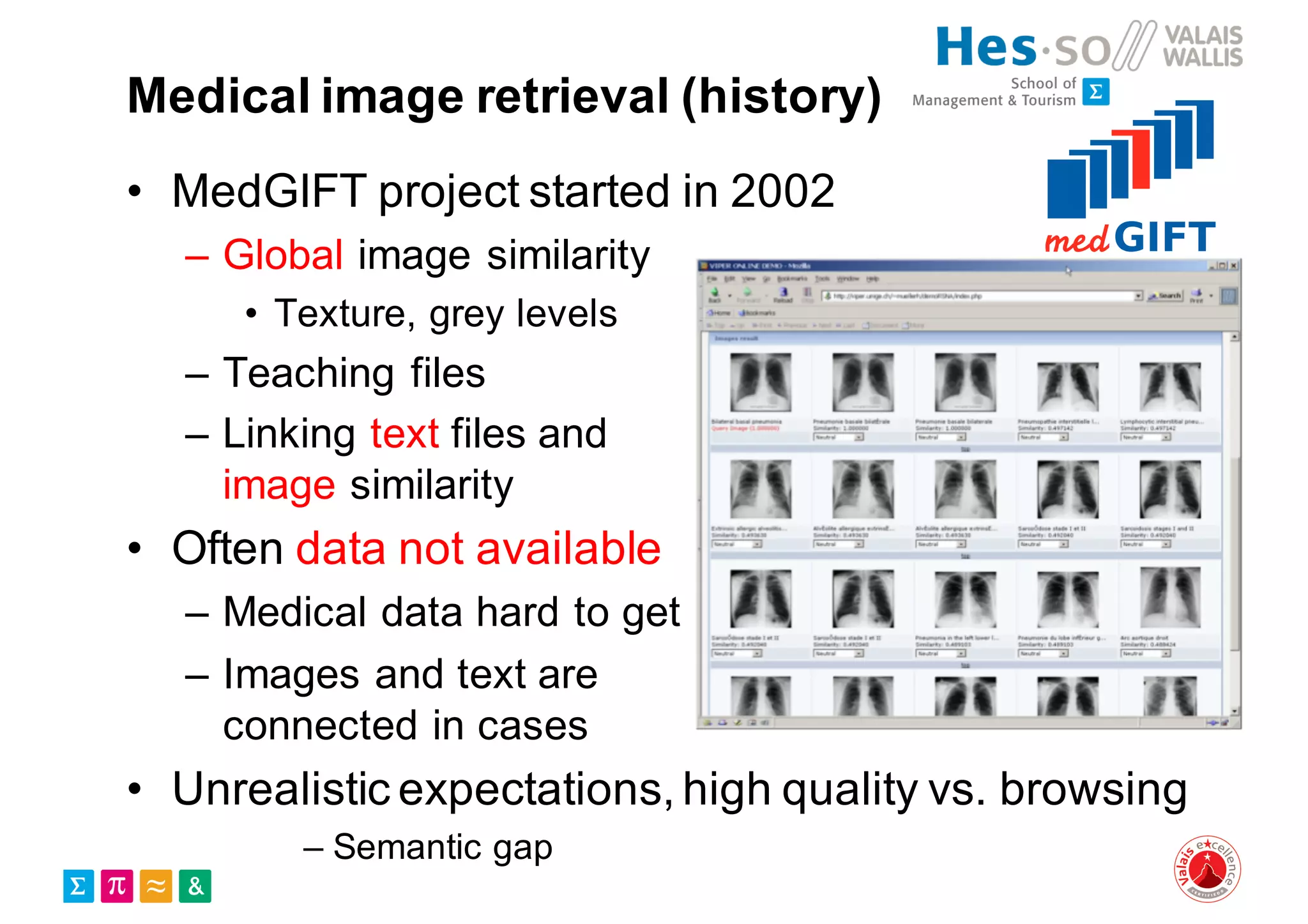
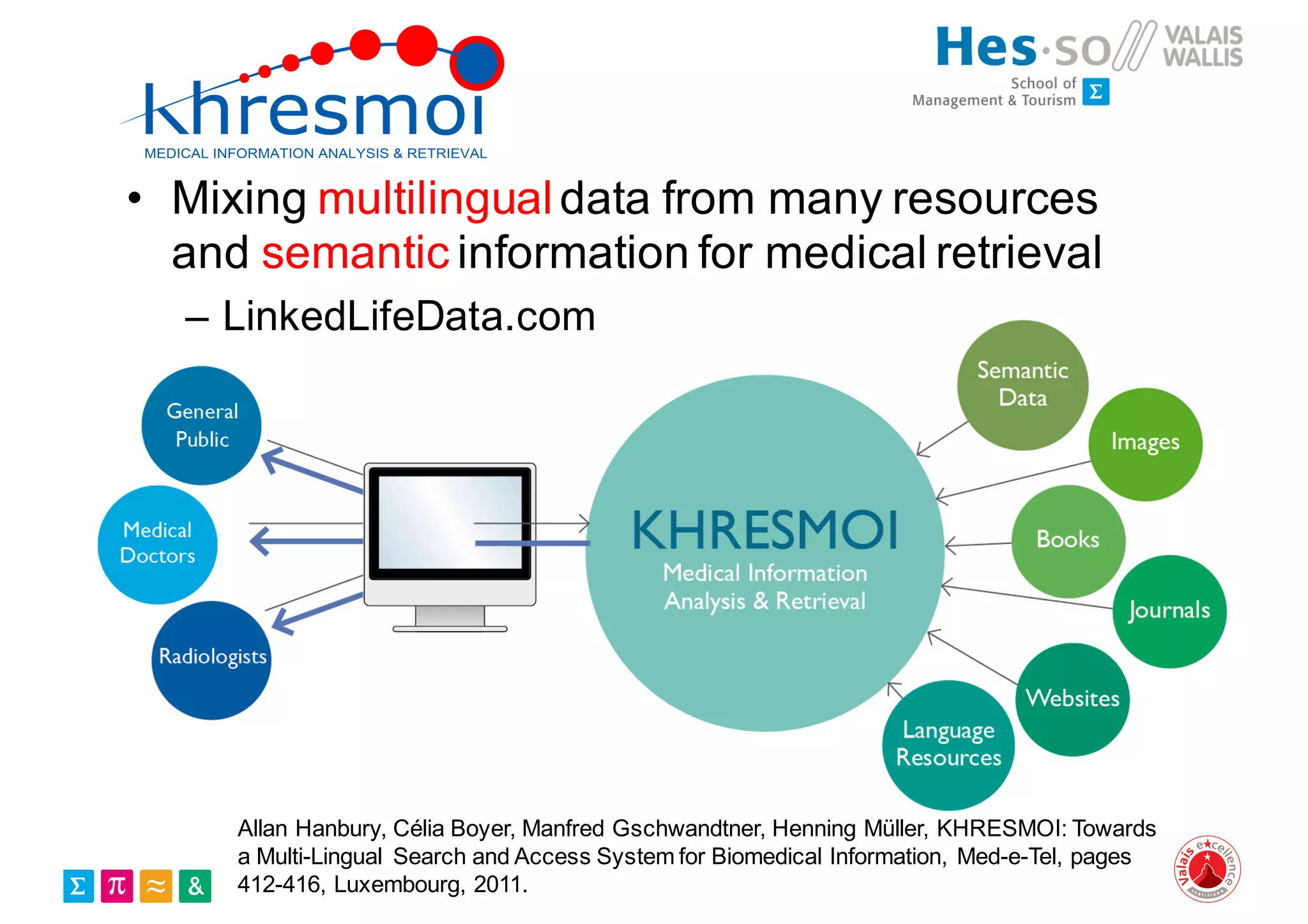
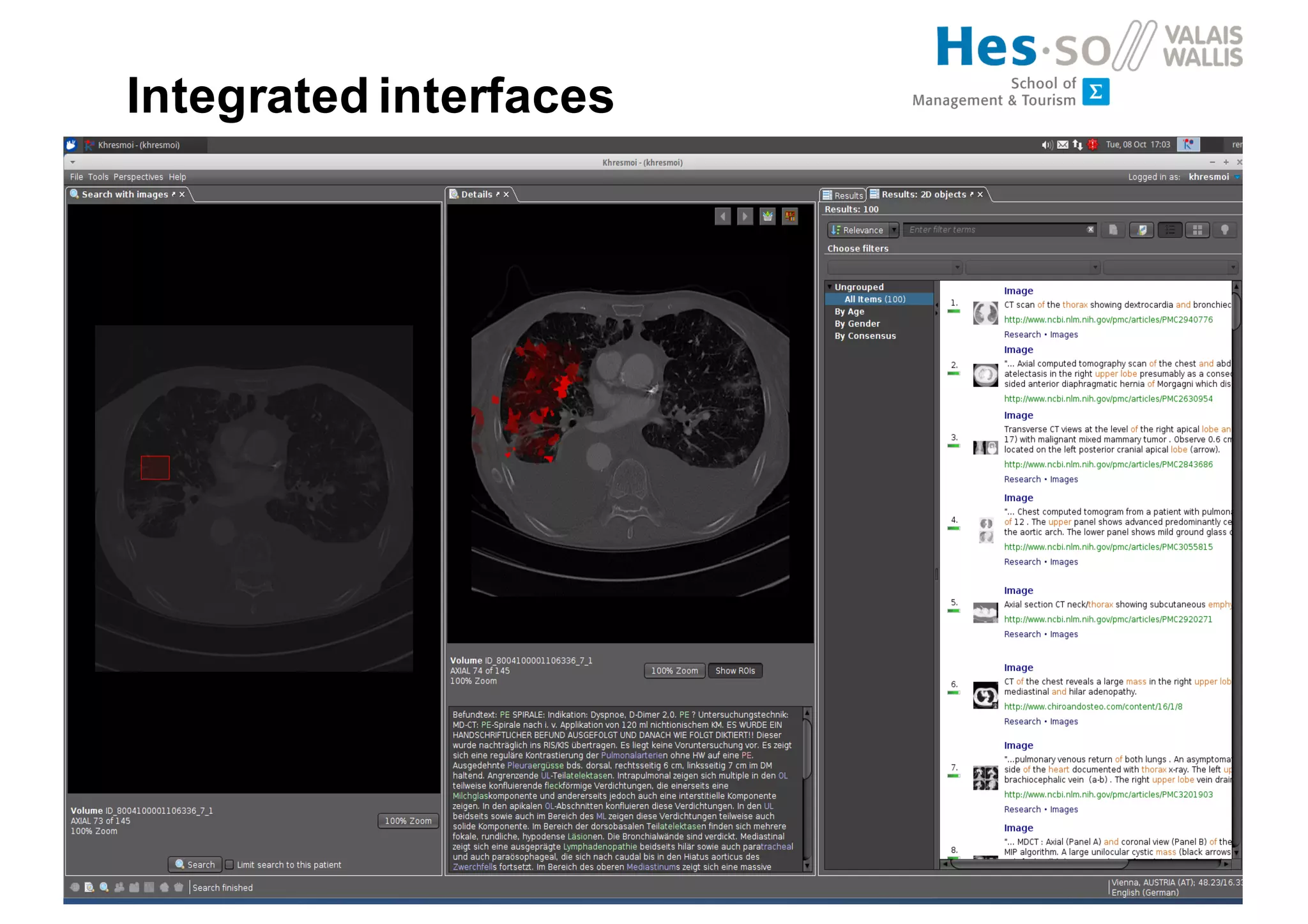
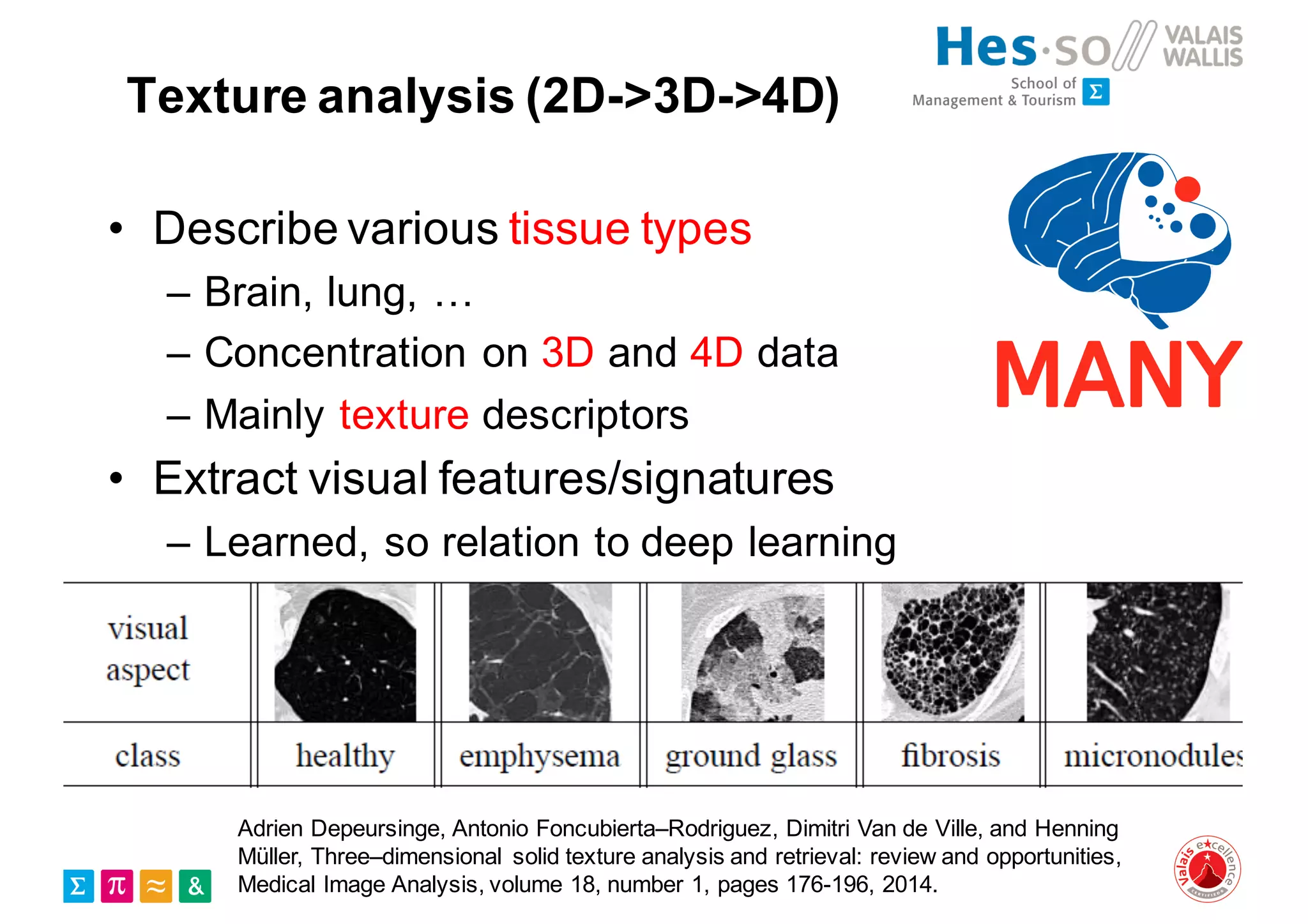
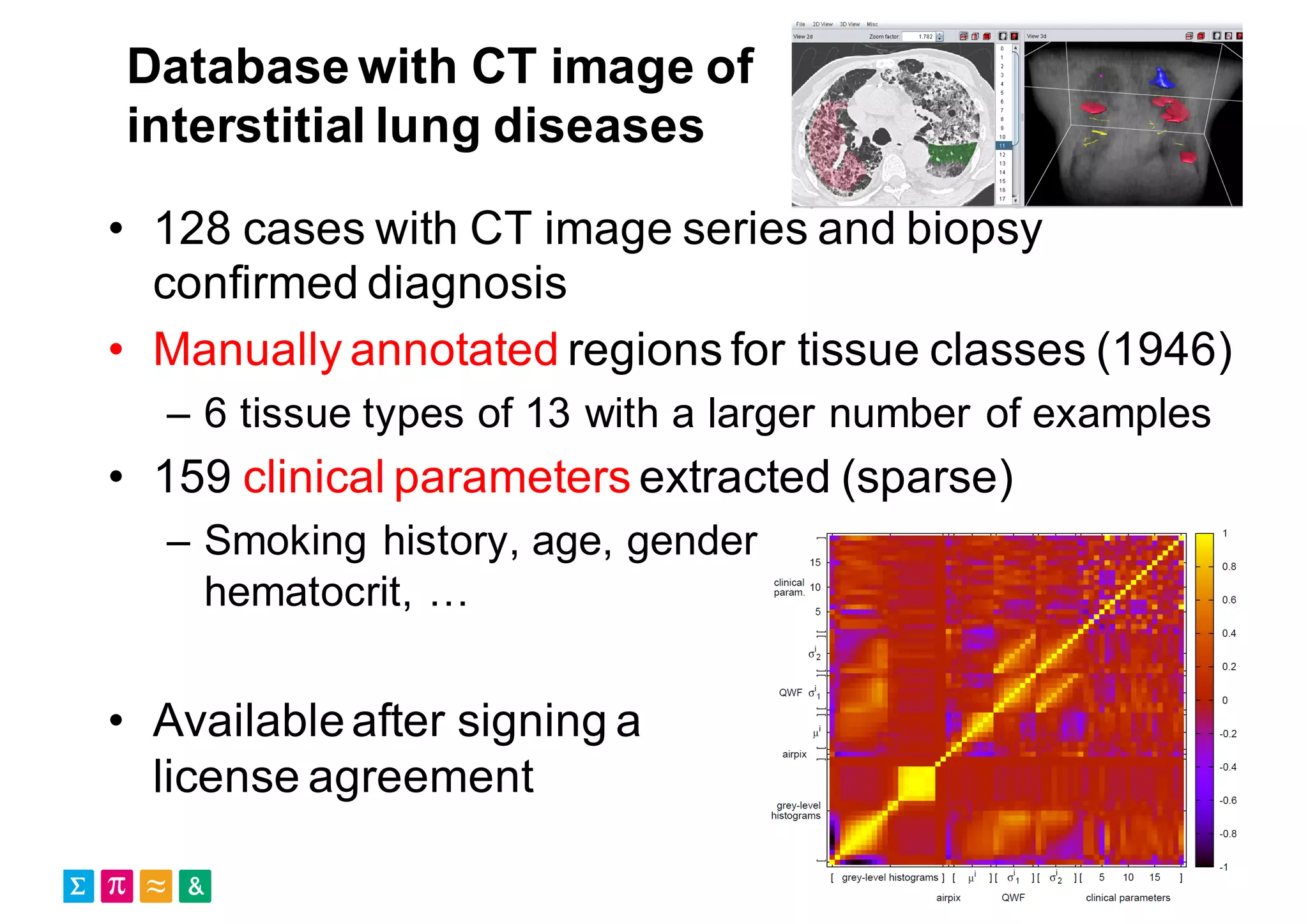
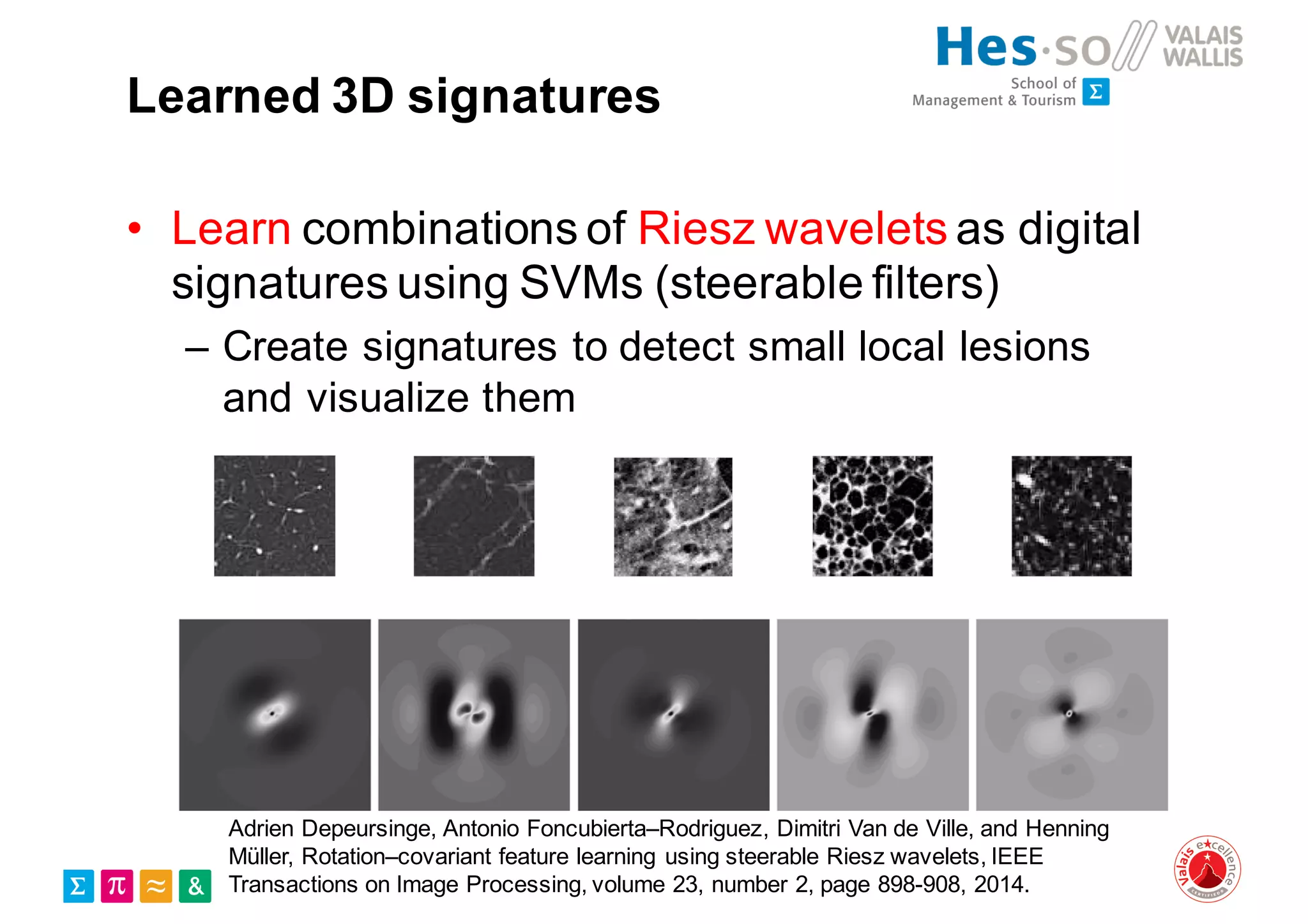
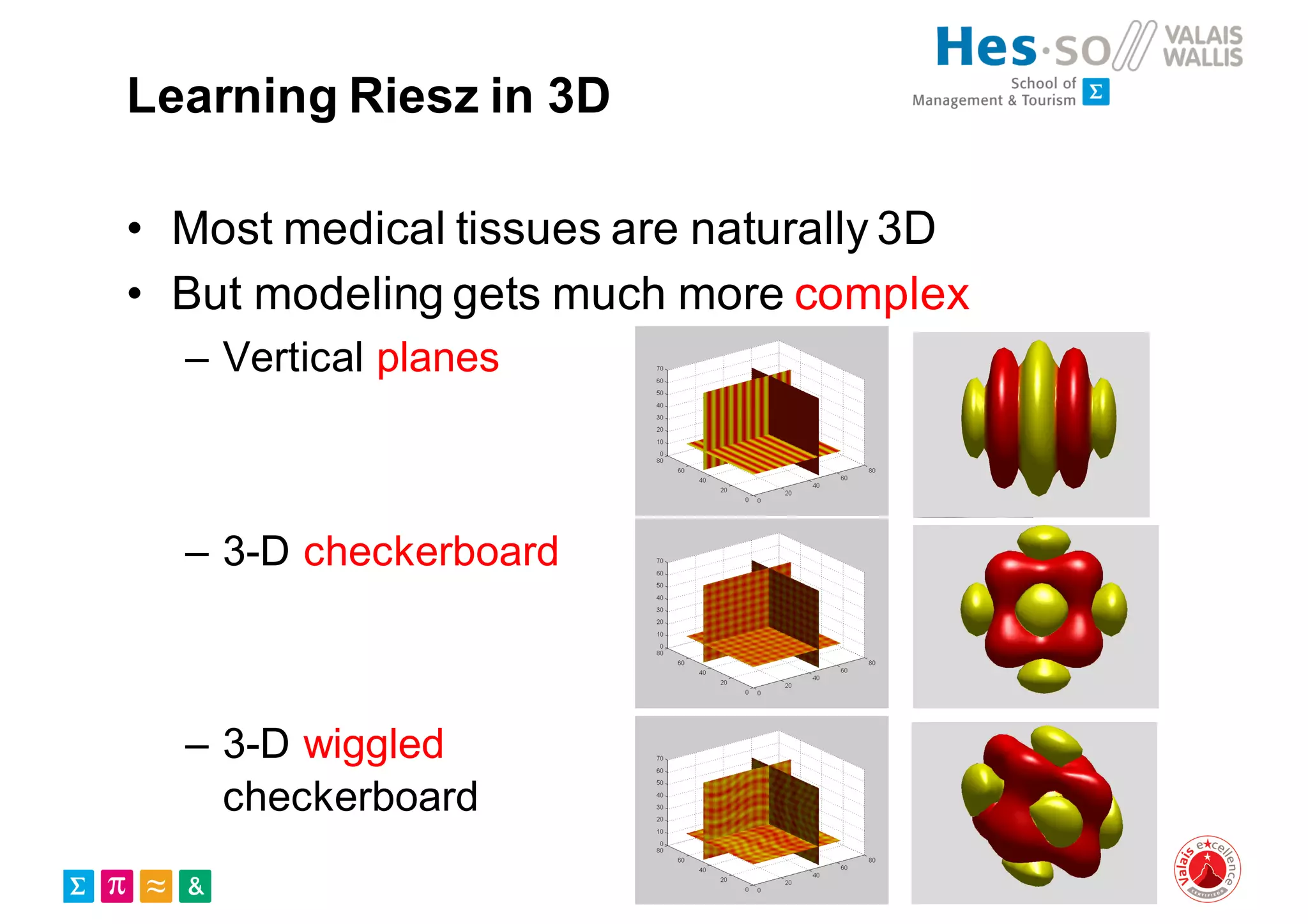
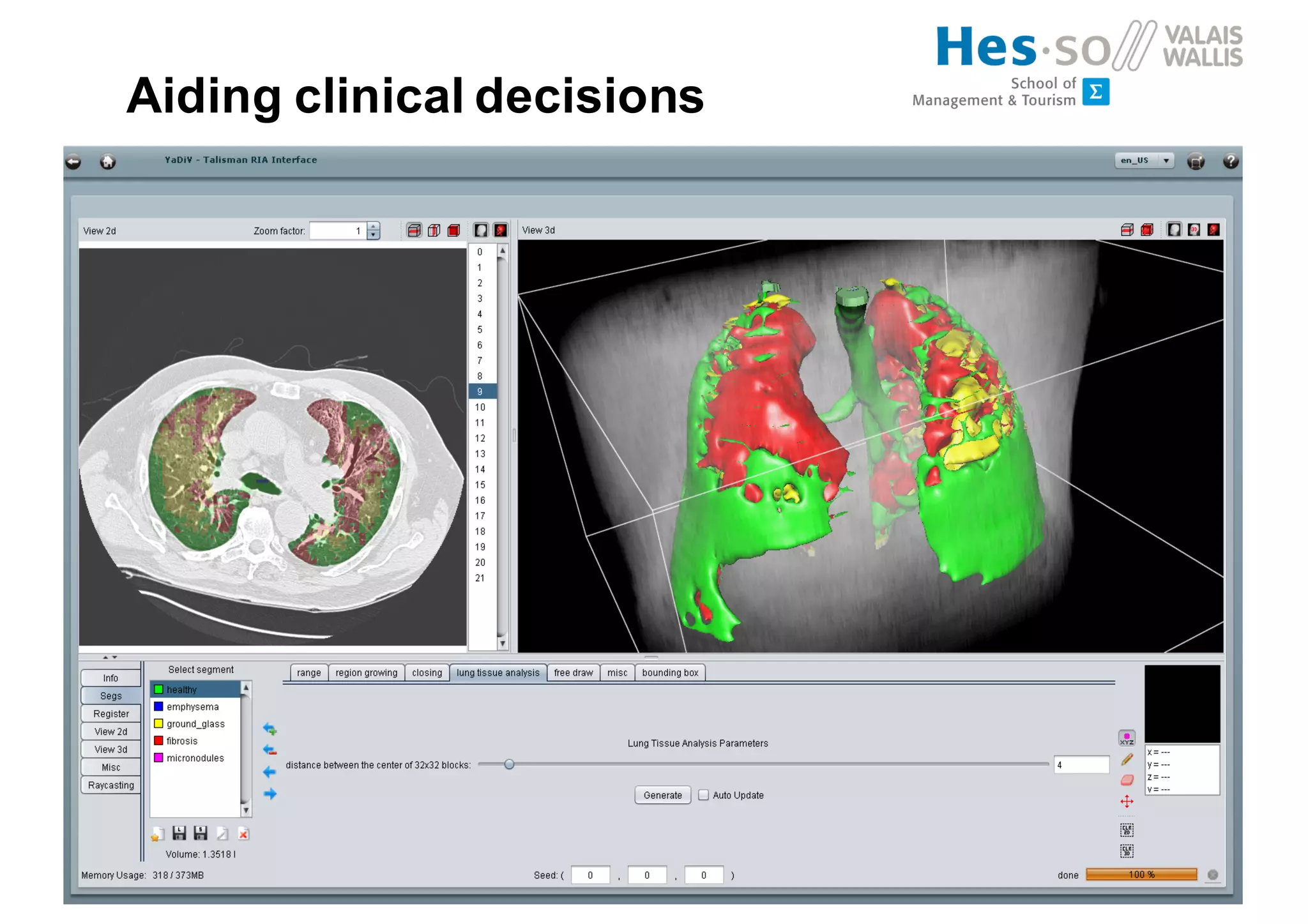
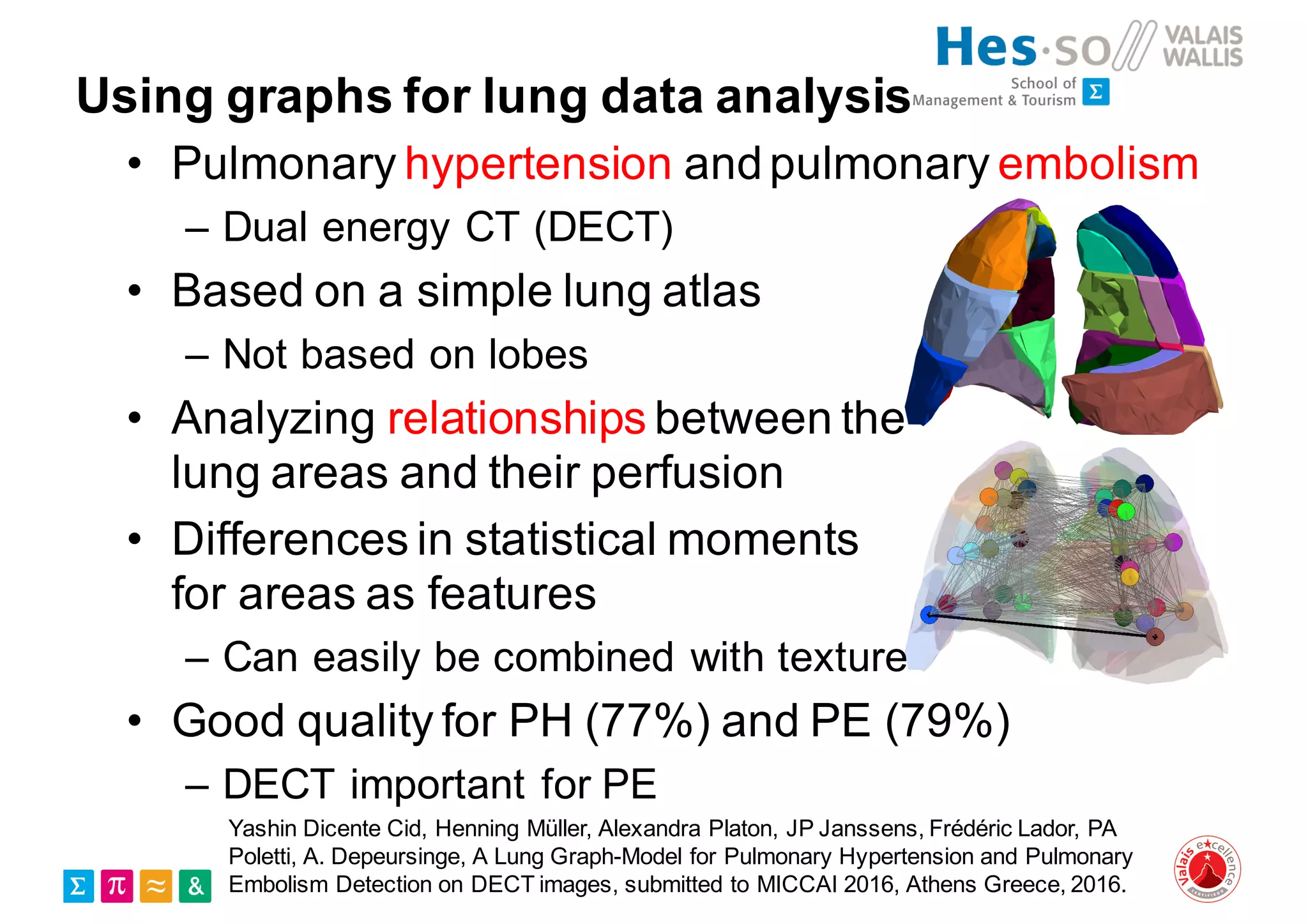
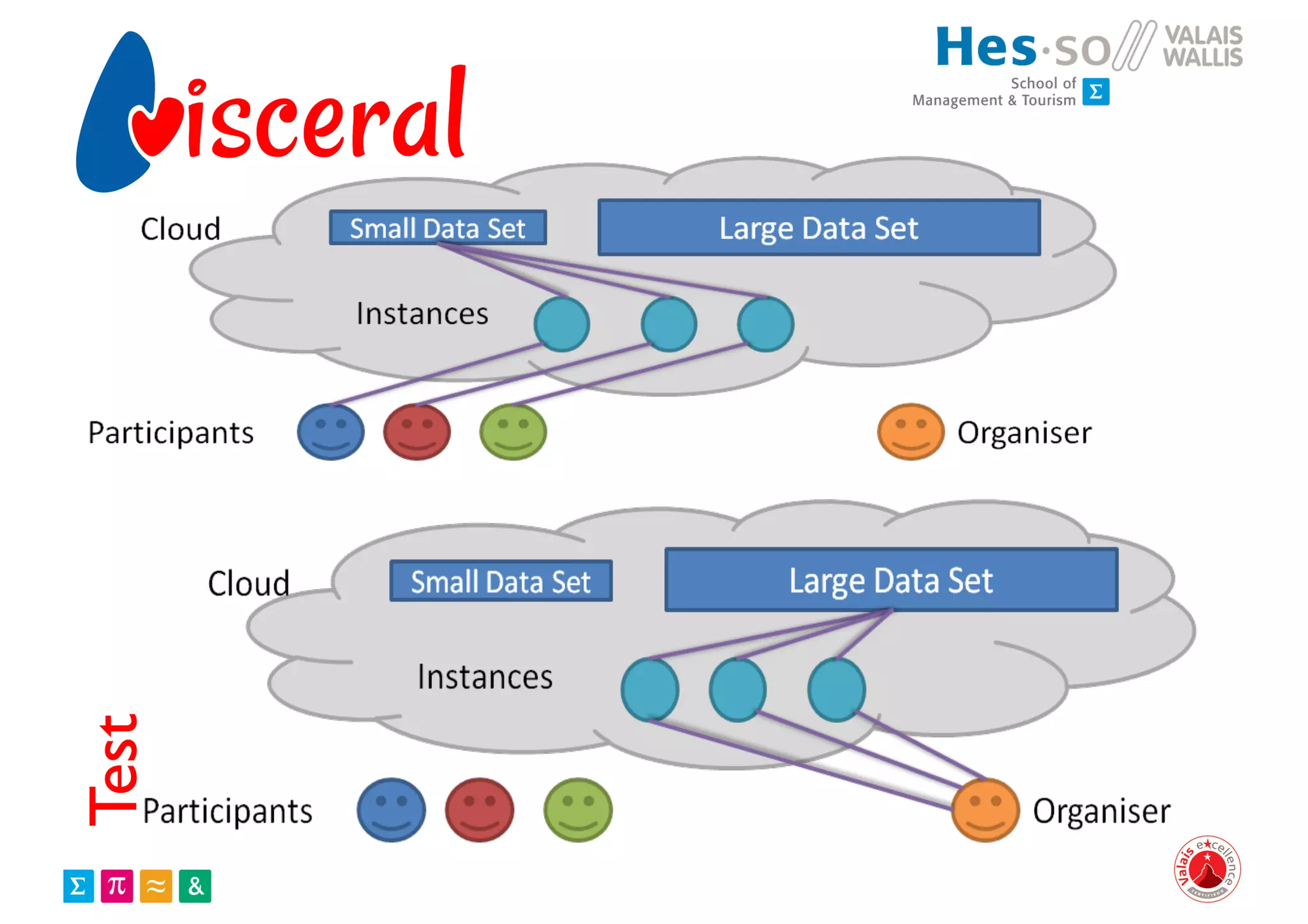
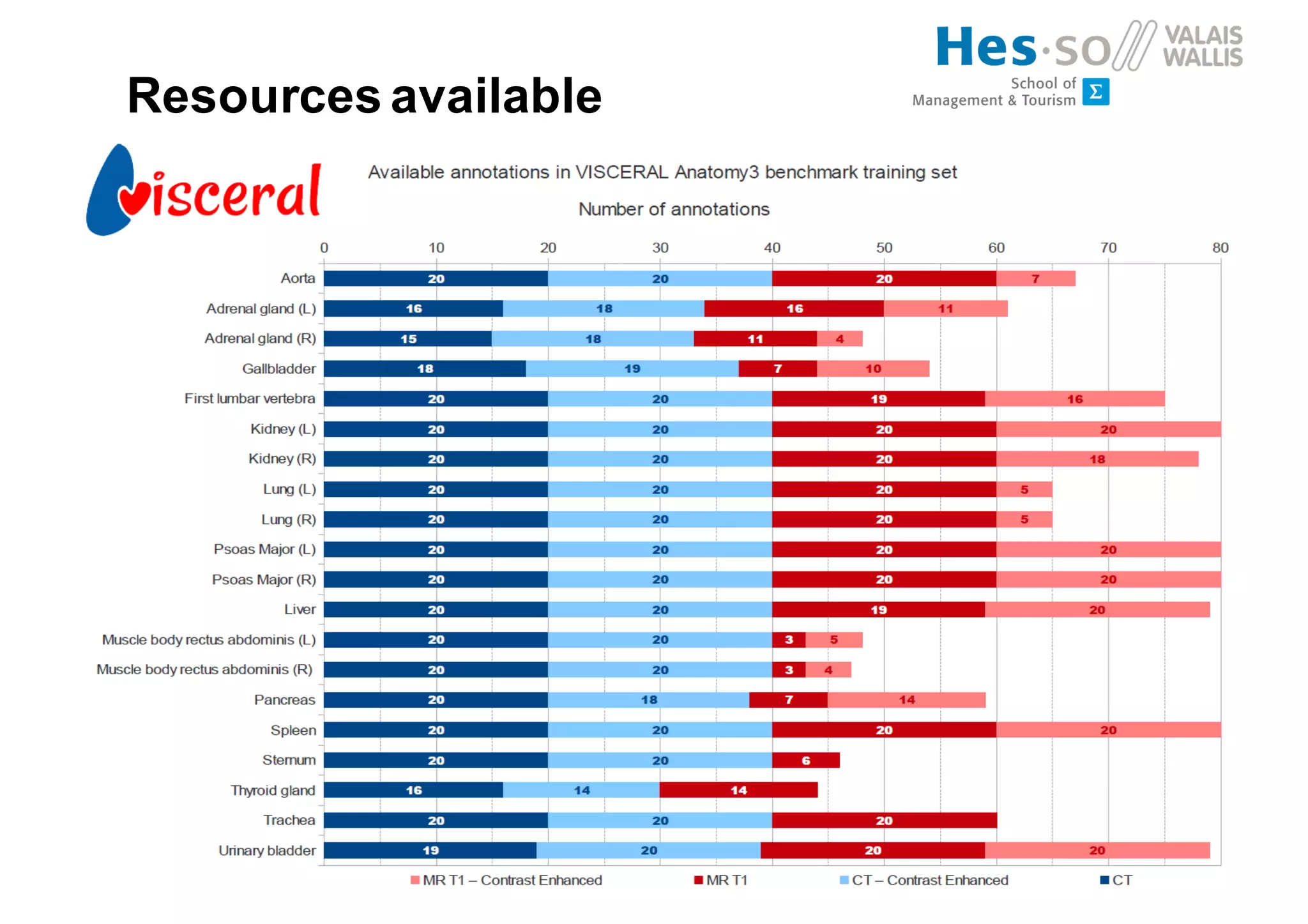
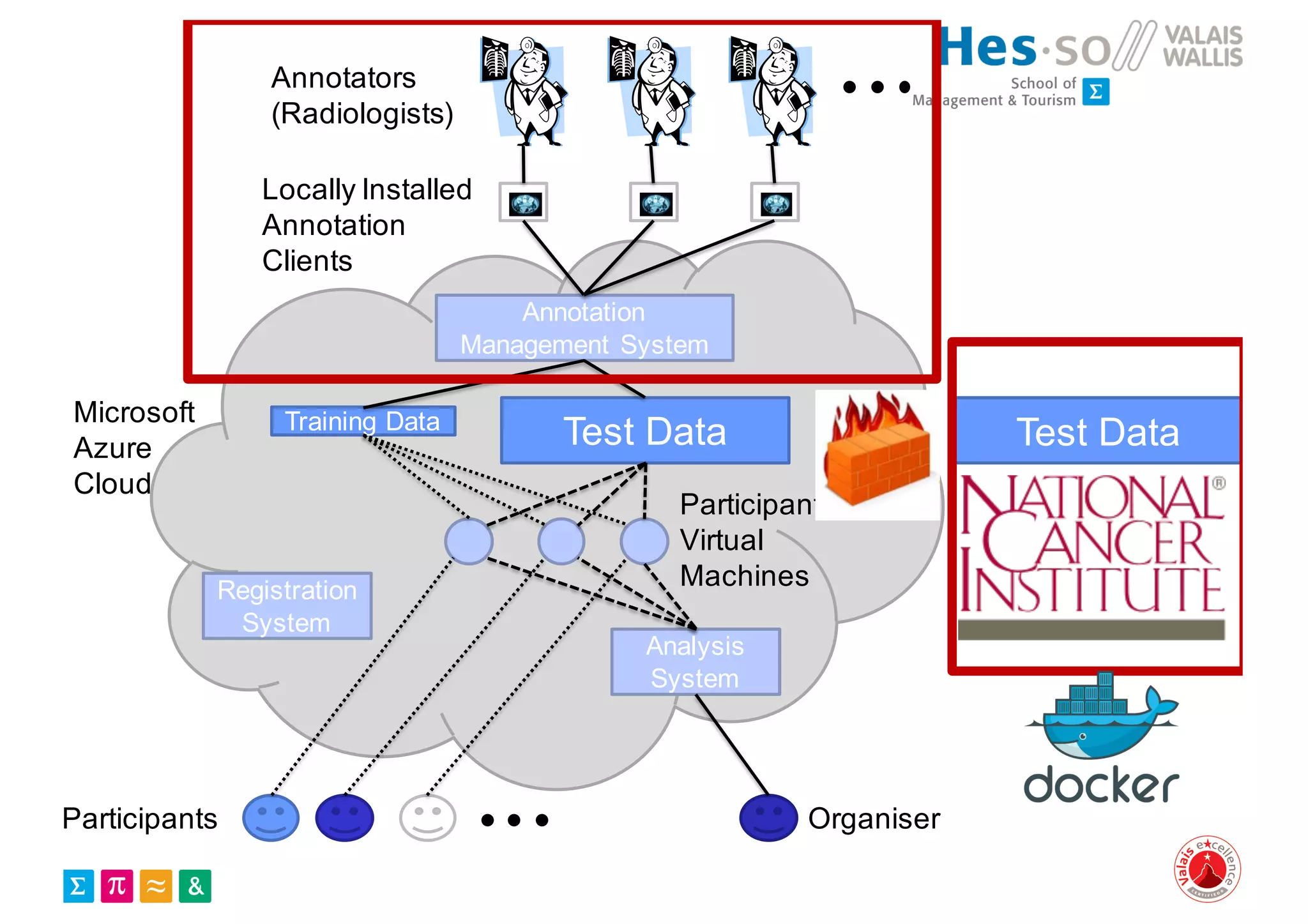
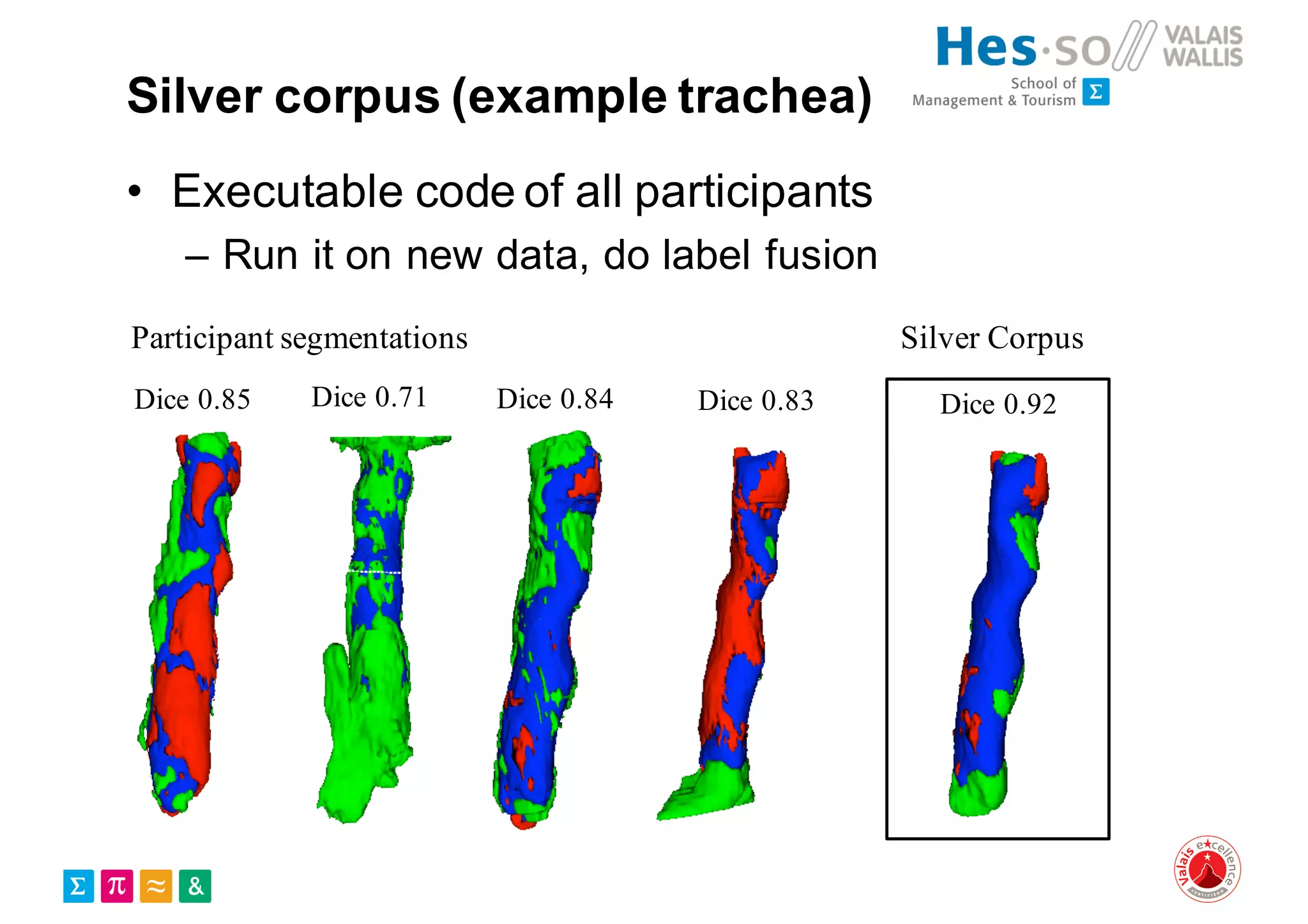
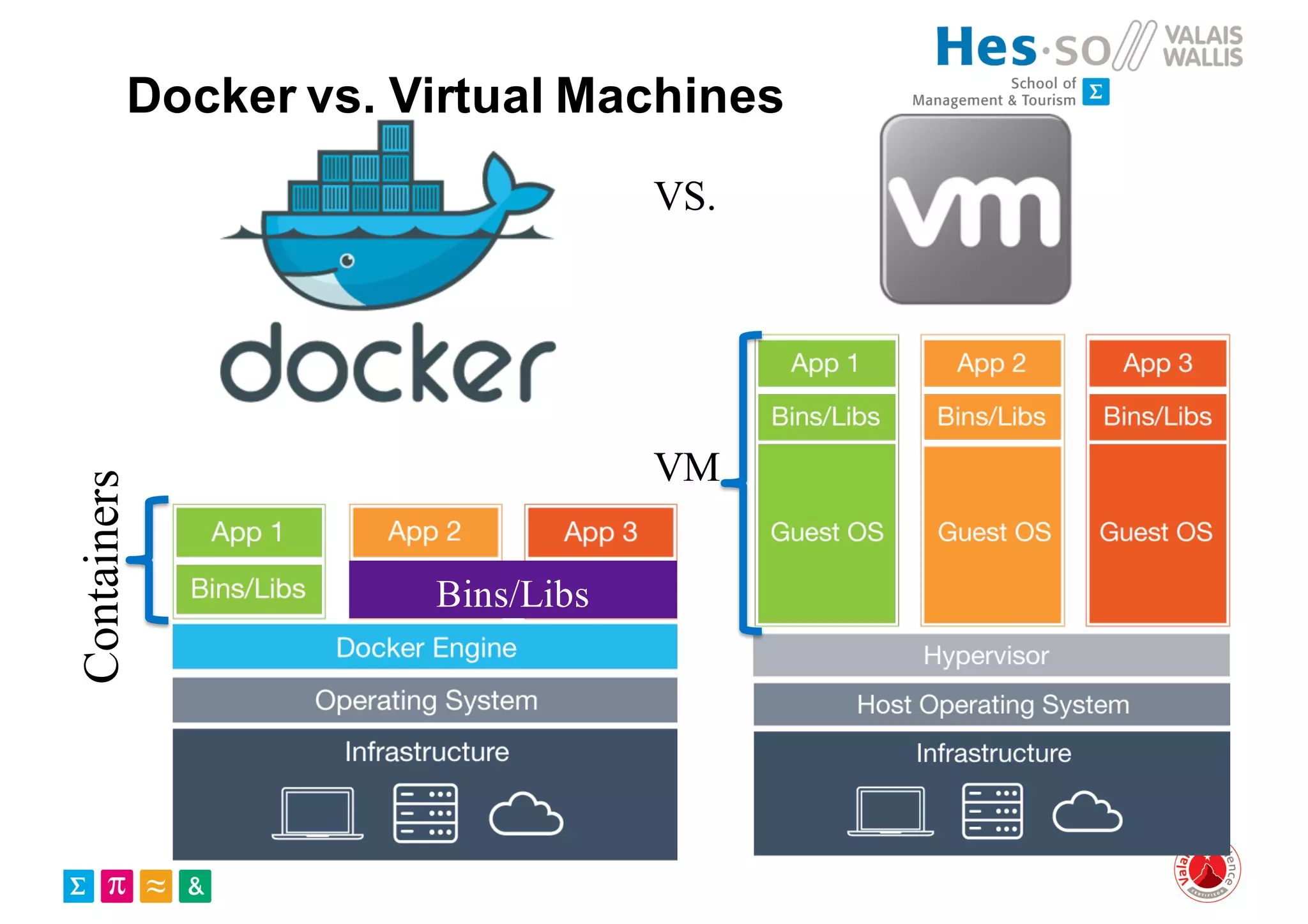
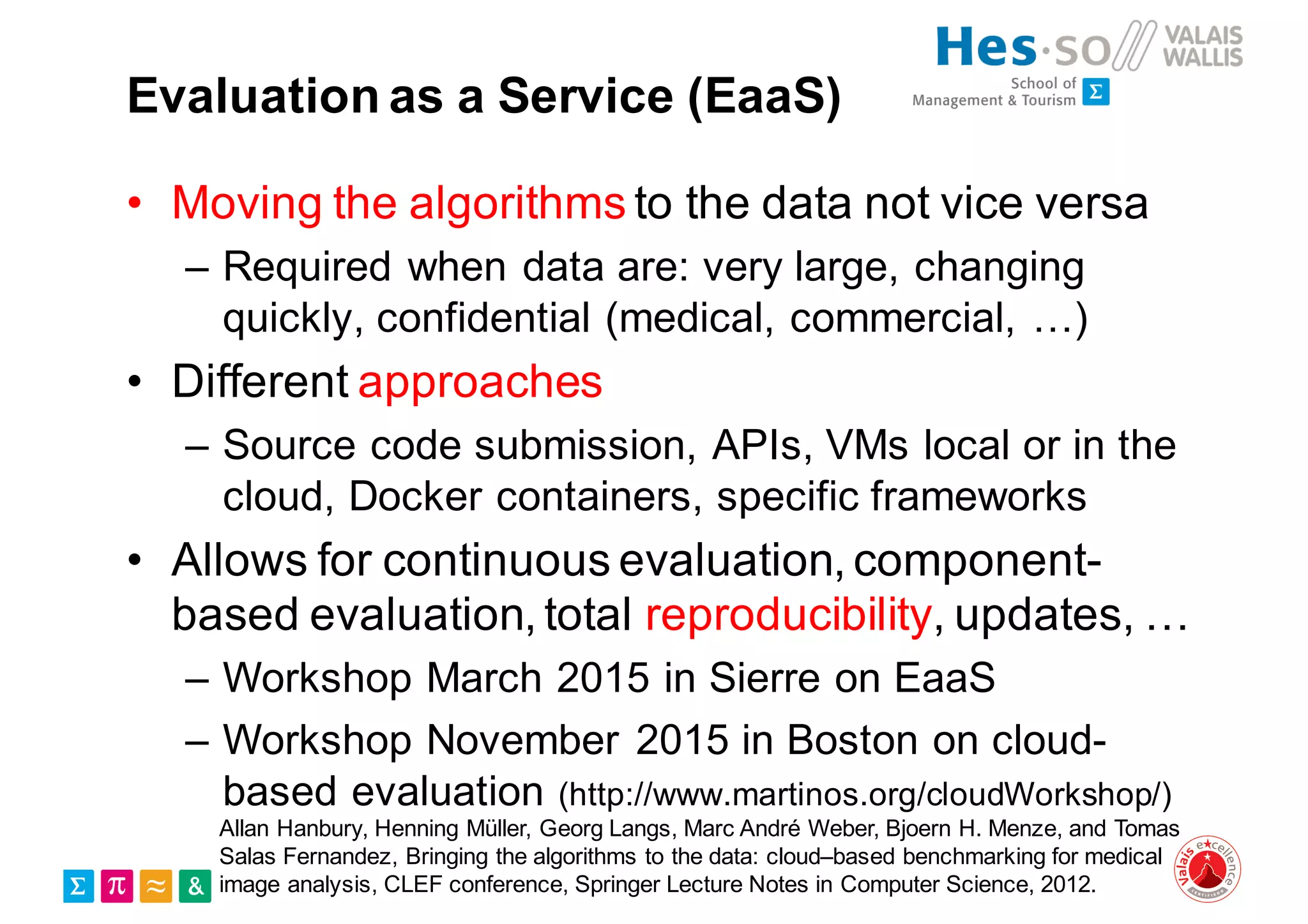
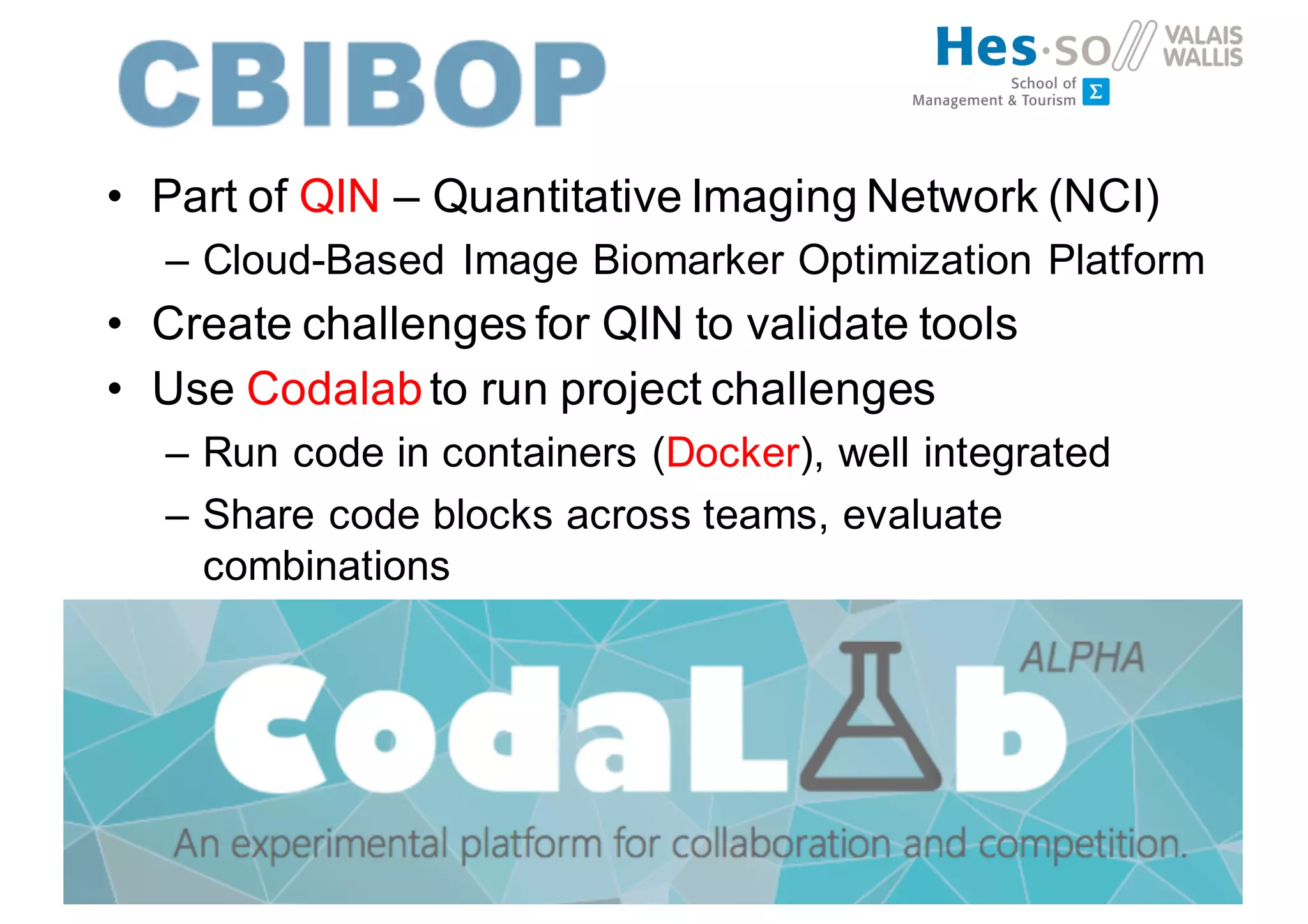
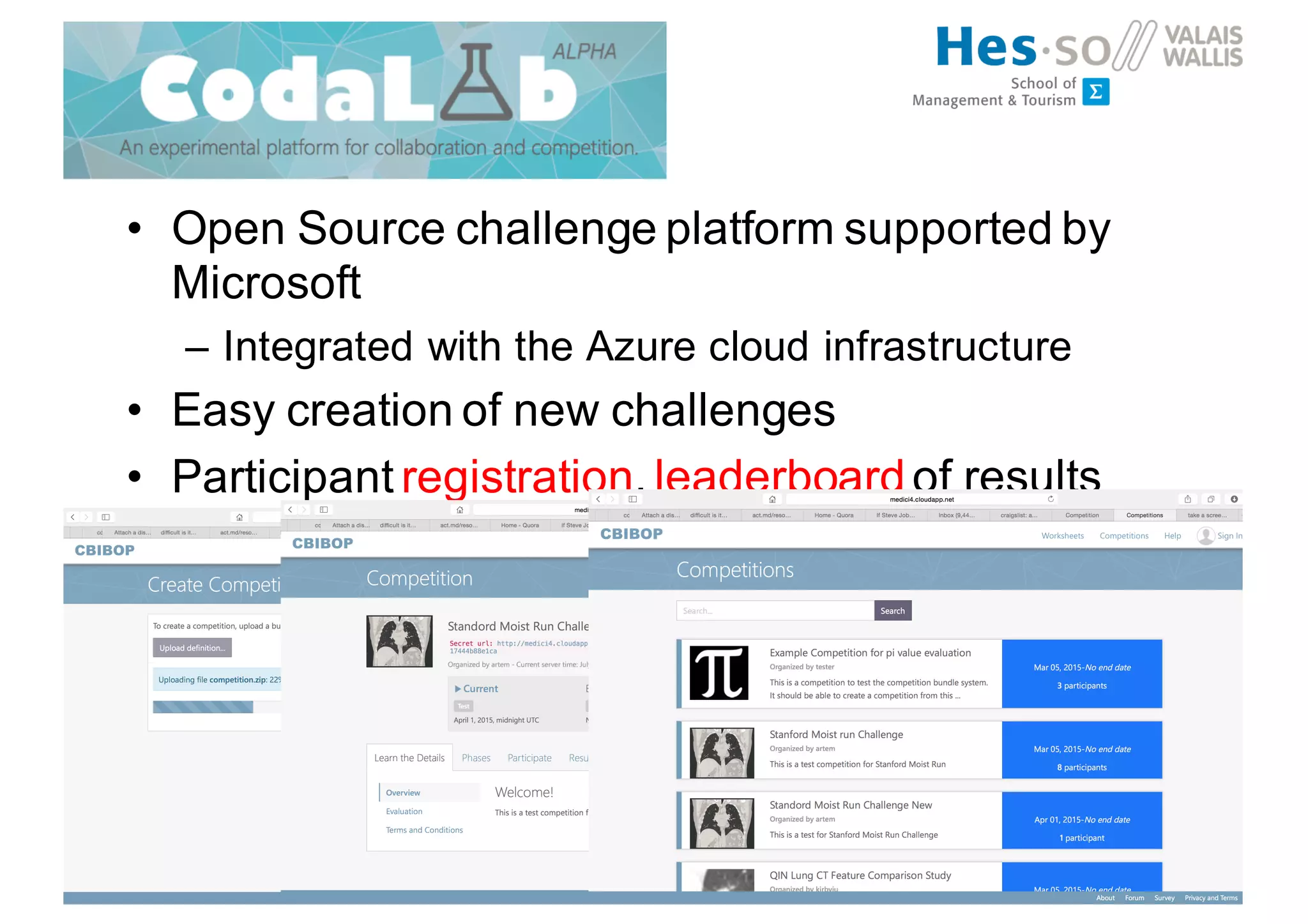
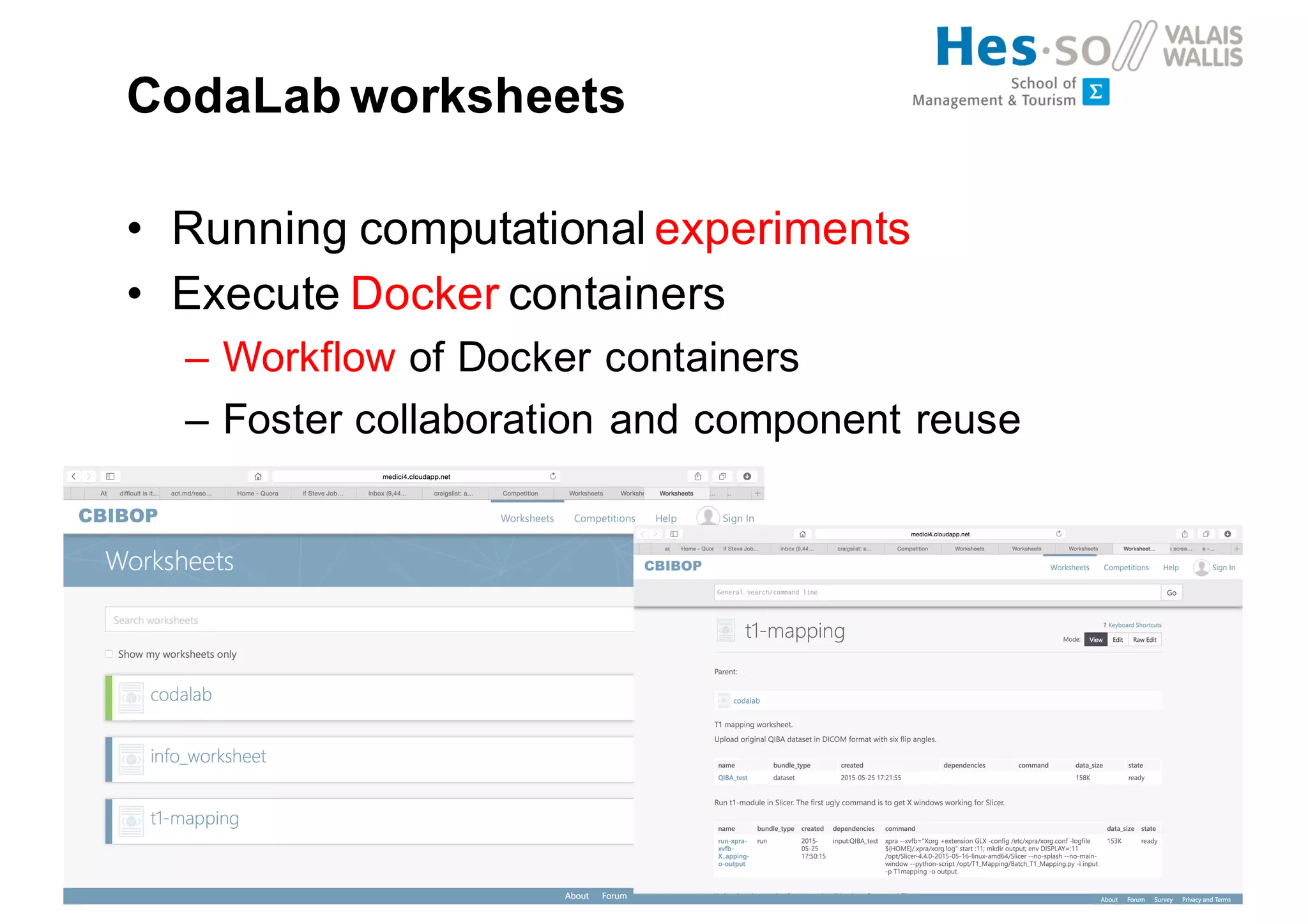
Henning Müller discusses the complexities of medical image analysis, big data evaluation, and the necessity for robust infrastructures to handle the vast and intricate imaging data utilized in medical diagnosis and treatment. The document outlines the history of medical image retrieval, highlighting challenges such as data availability and the need for effective analysis tools, while also exploring future directions including cloud computing and crowdsourcing in research. Key projects and evidence-based strategies are presented to enhance image retrieval and ensure that medical data remains accessible and usable.