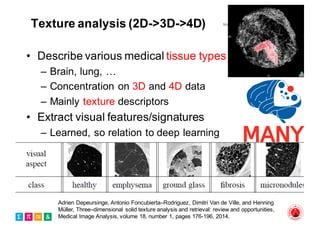
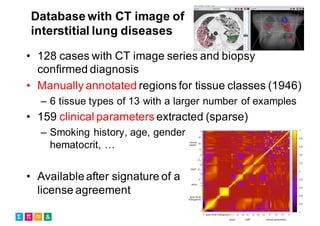
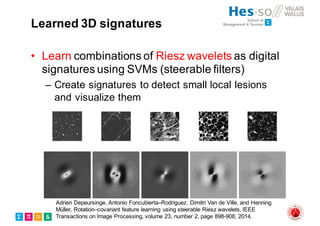
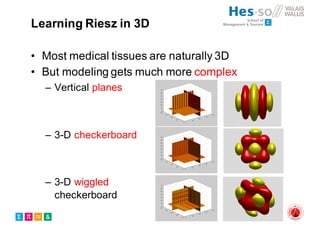
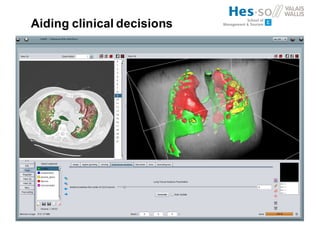
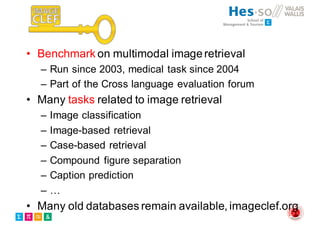
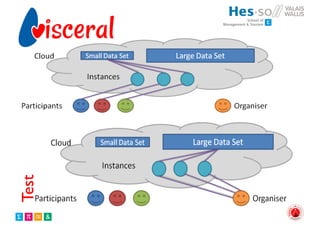
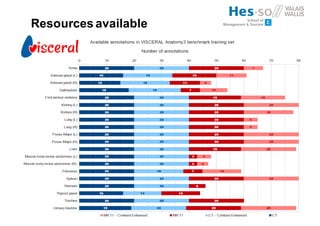
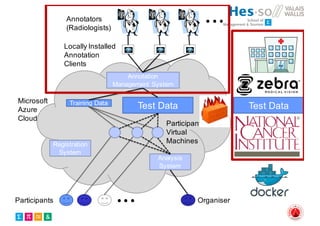
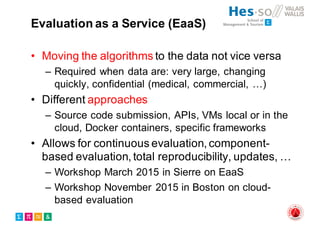
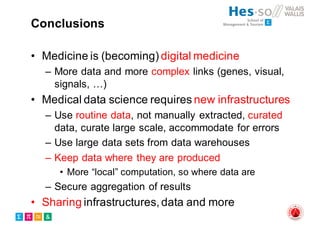
The document outlines the evolution and current state of medical image analysis, retrieval, and evaluation infrastructures, highlighting key projects and challenges in accessing medical imaging data. It emphasizes the importance of data-sharing, quality evaluation of algorithms, and integrating various data sources in the field of medical informatics. Additionally, it discusses future directions aimed at improving data accessibility and research efficiency through advanced computational methods and collaborative efforts.