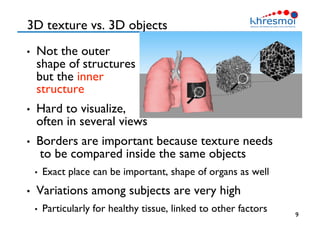
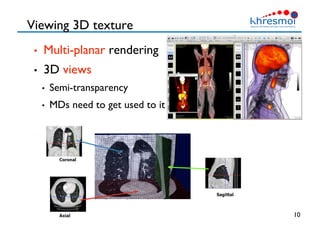
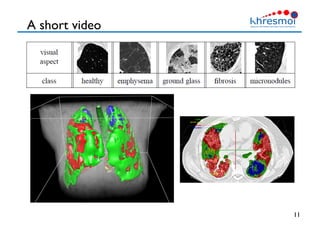
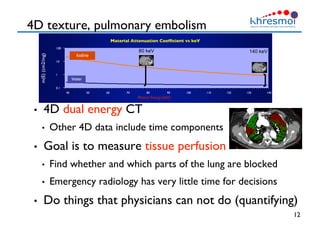
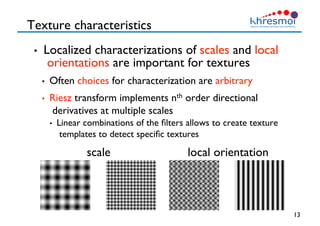
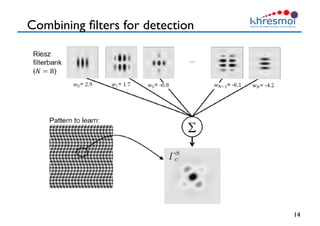
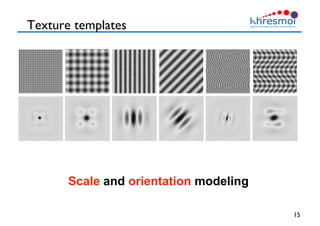
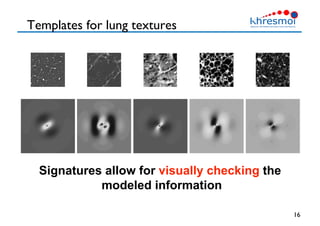
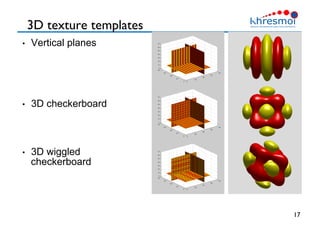
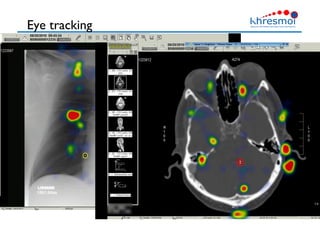
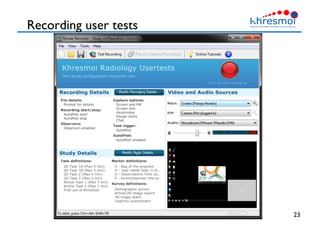
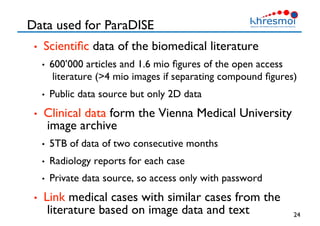
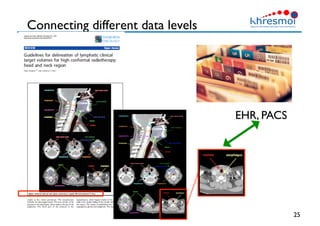
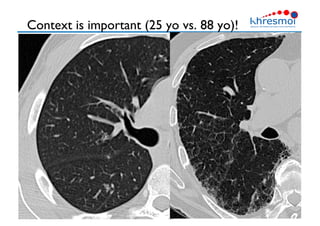
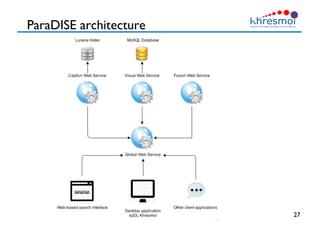
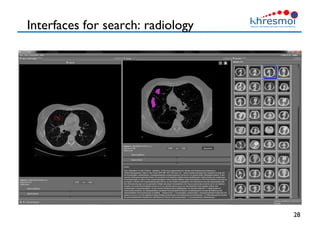
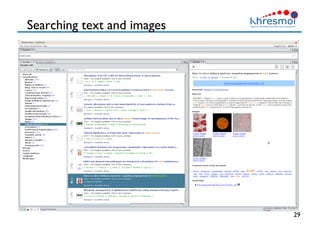
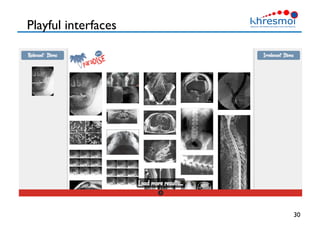
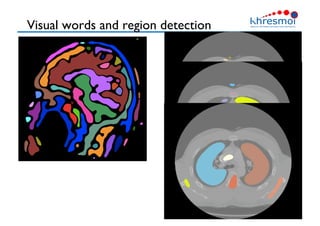
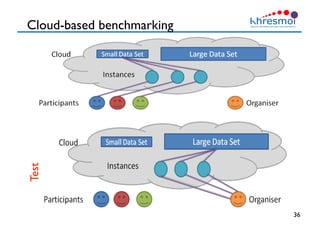
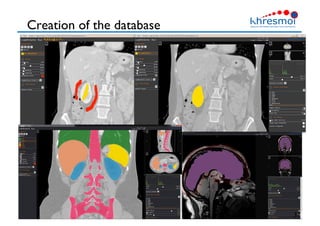
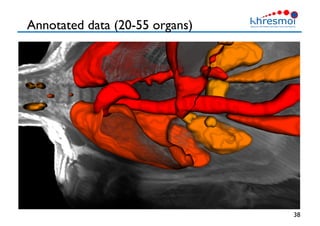
The document provides an overview of Henning Müller's background in medical informatics and his work in medical 3D data retrieval, emphasizing the increasing complexity of imaging data in diagnostics. It discusses various topics such as 3D texture analysis, the necessity of clinical data for accurate diagnoses, and the challenges faced in radiology image retrieval and organ detection. The document also outlines ongoing projects aimed at improving systems for analyzing and retrieving medical imaging information, with a focus on the importance of context and collaborative efforts in the field.