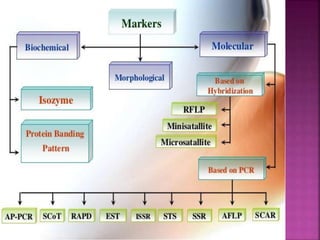
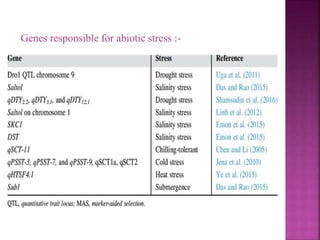
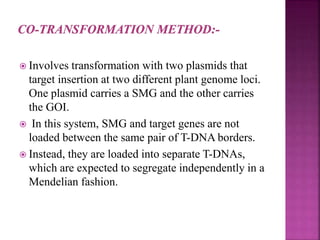
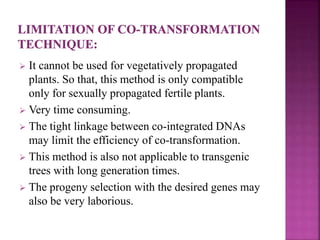
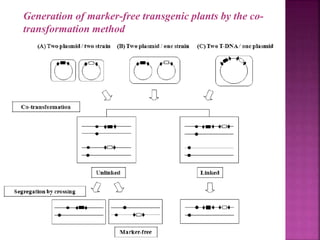
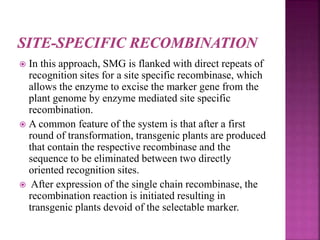
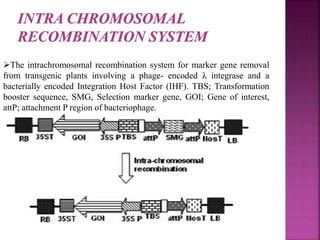
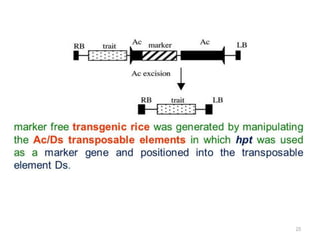
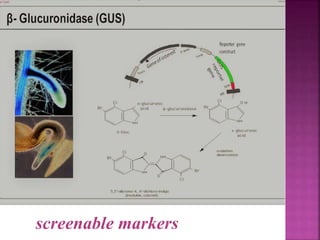
The document discusses career opportunities in biotechnology, focusing on the development of marker-free transgenic plants and the various strategies employed to eliminate marker genes traditionally used in plant transformation. It examines methods such as positive and negative selection systems, co-transformation, and site-specific recombination to achieve this goal while addressing biosafety concerns and the ecological implications of genetic engineering. The overall aim is to enhance public acceptance of transgenic plants by ensuring they do not carry selectable marker genes that may pose risks to the environment and food safety.