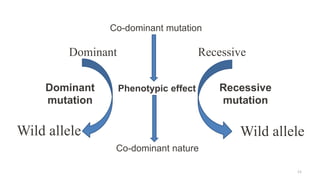
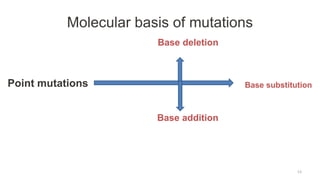
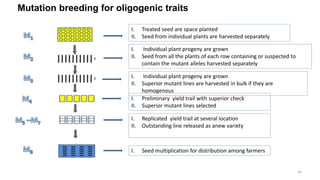
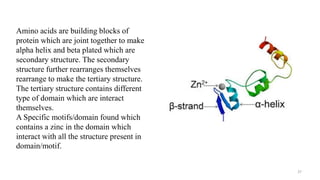
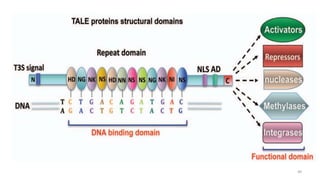
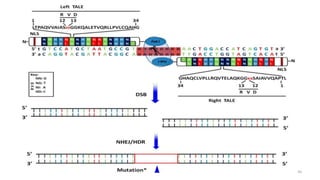
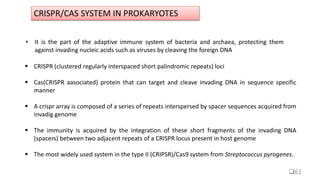
Targeted mutagenesis is a molecular biology technique used to intentionally induce specific changes to the DNA sequence of a gene. Zinc finger nucleases (ZFNs) are one method for targeted mutagenesis. ZFNs consist of a DNA-binding domain containing zinc finger motifs and a DNA-cleavage domain from the FokI restriction enzyme. The zinc finger domain binds to a specific DNA sequence while the FokI domain cleaves the DNA, creating a double-strand break. This double-strand break stimulates the cell's natural DNA repair processes and can result in targeted gene deletions, insertions or modifications.