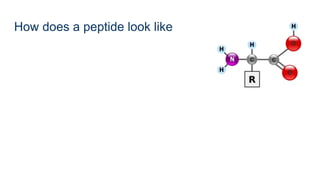
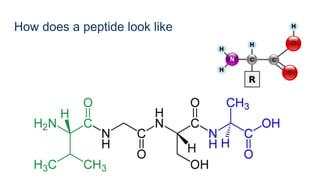
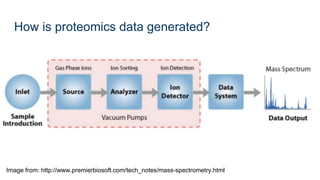
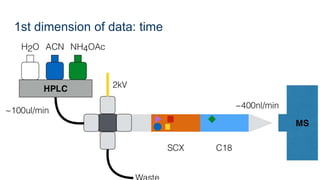
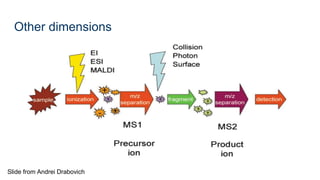
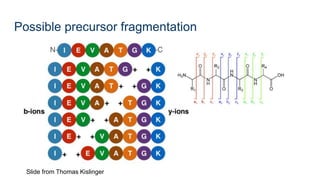
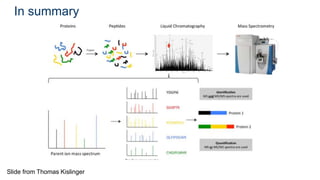
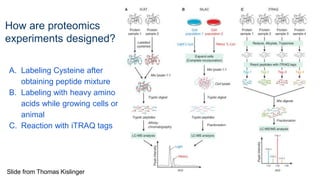
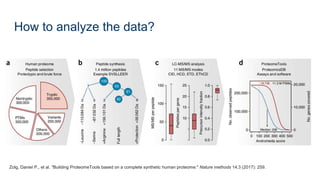
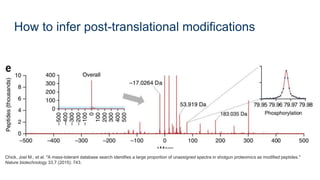
This document provides an introduction to proteomics. It discusses why proteomics is important as proteins carry out most biological functions. It also mentions valuable publicly available proteomics resources and applications of machine learning in proteomics data analysis and interpretation. The document describes how proteomics data is generated using trypsin digestion and mass spectrometry techniques. It discusses the different dimensions of proteomics data and how experiments are designed using labeling or tagging approaches. Finally, it outlines the features of proteomic data, approaches to data analysis including de novo sequencing and identifying post-translational modifications, relevant bioinformatics tools and publicly available cancer proteomics resources.