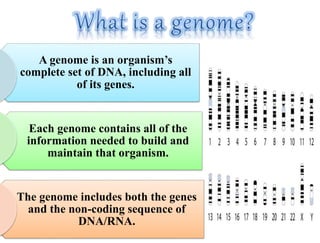
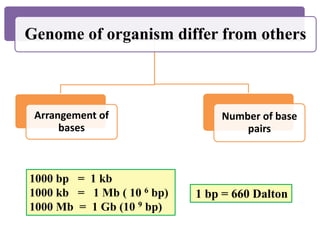
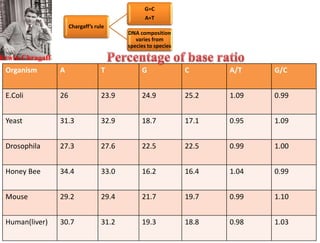
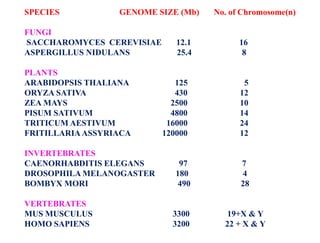
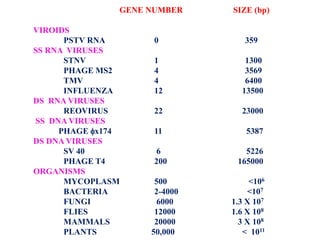
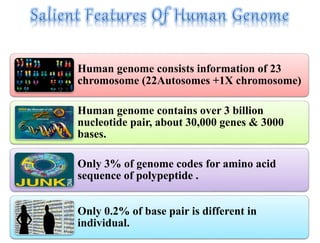
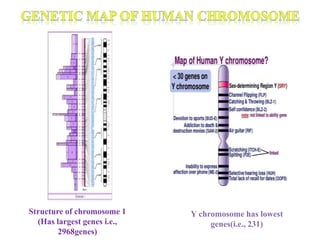
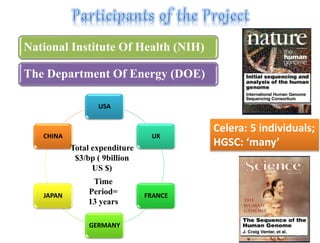
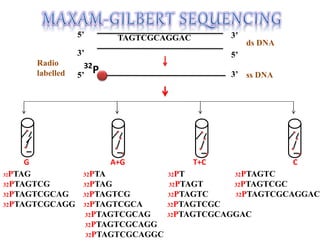
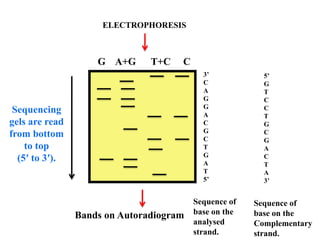
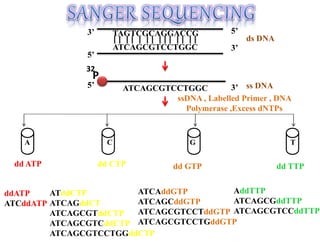
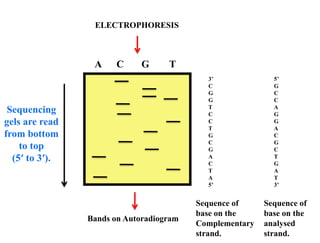
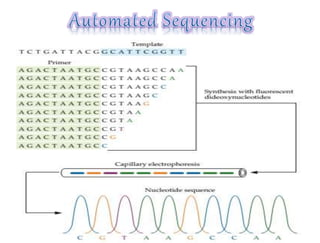
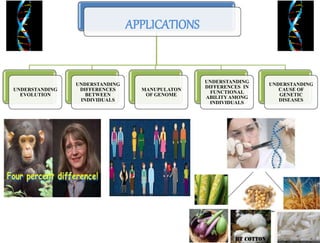
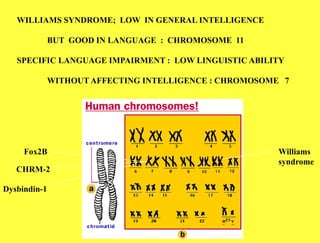
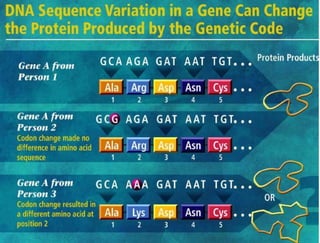
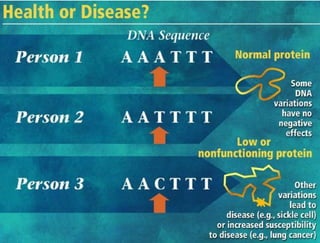
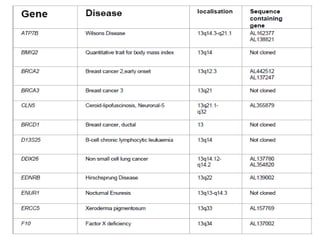
This document summarizes key aspects of the human genome project including its goals, methods of DNA sequencing, applications, and ongoing challenges. The human genome project was an international scientific research project that determined the sequence of nucleotide base pairs that make up human DNA and mapped the approximate locations of genes on each of the 23 chromosome pairs. It aimed to identify all the genes in human DNA, determine the sequences of the 3 billion chemical base pairs that make up human DNA, store this information in databases, improve tools for data analysis, and transfer related technologies to the private sector.