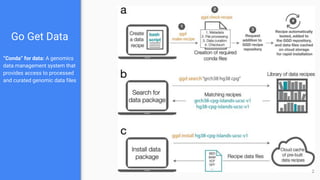
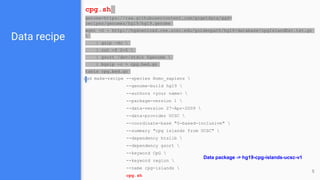
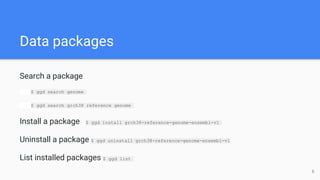
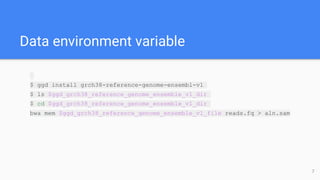
Go Get Data (GGD) is a genomics data management system that provides access to processed and curated genomic data files. It allows users to create "data recipes" that define genomic data files and their metadata. These recipes are used to generate data packages that can be installed and their files accessed via environment variables. GGD also supports finding, installing, uninstalling, and listing installed data packages.