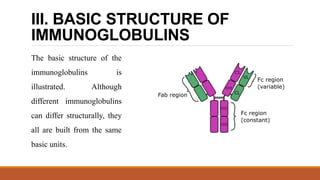
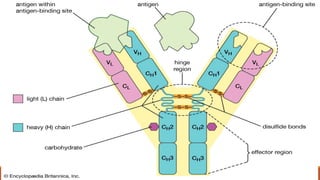
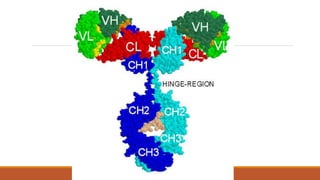
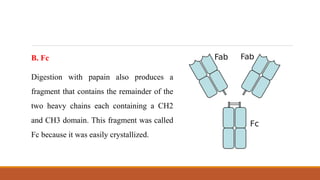
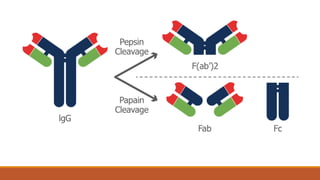
Immunoglobulins are glycoprotein molecules composed of two light chains and two heavy chains that function as antibodies. They bind specifically to antigens through complementarity determining regions in their variable regions. This binding can result in various effector functions like complement activation or binding to immune cells. Immunoglobulin molecules can be cleaved by enzymes into fragments that isolate different functional properties - Fab fragments contain the antigen binding site, Fc fragments mediate effector functions, and F(ab')2 fragments are divalent for antigen binding but lack effector functions.