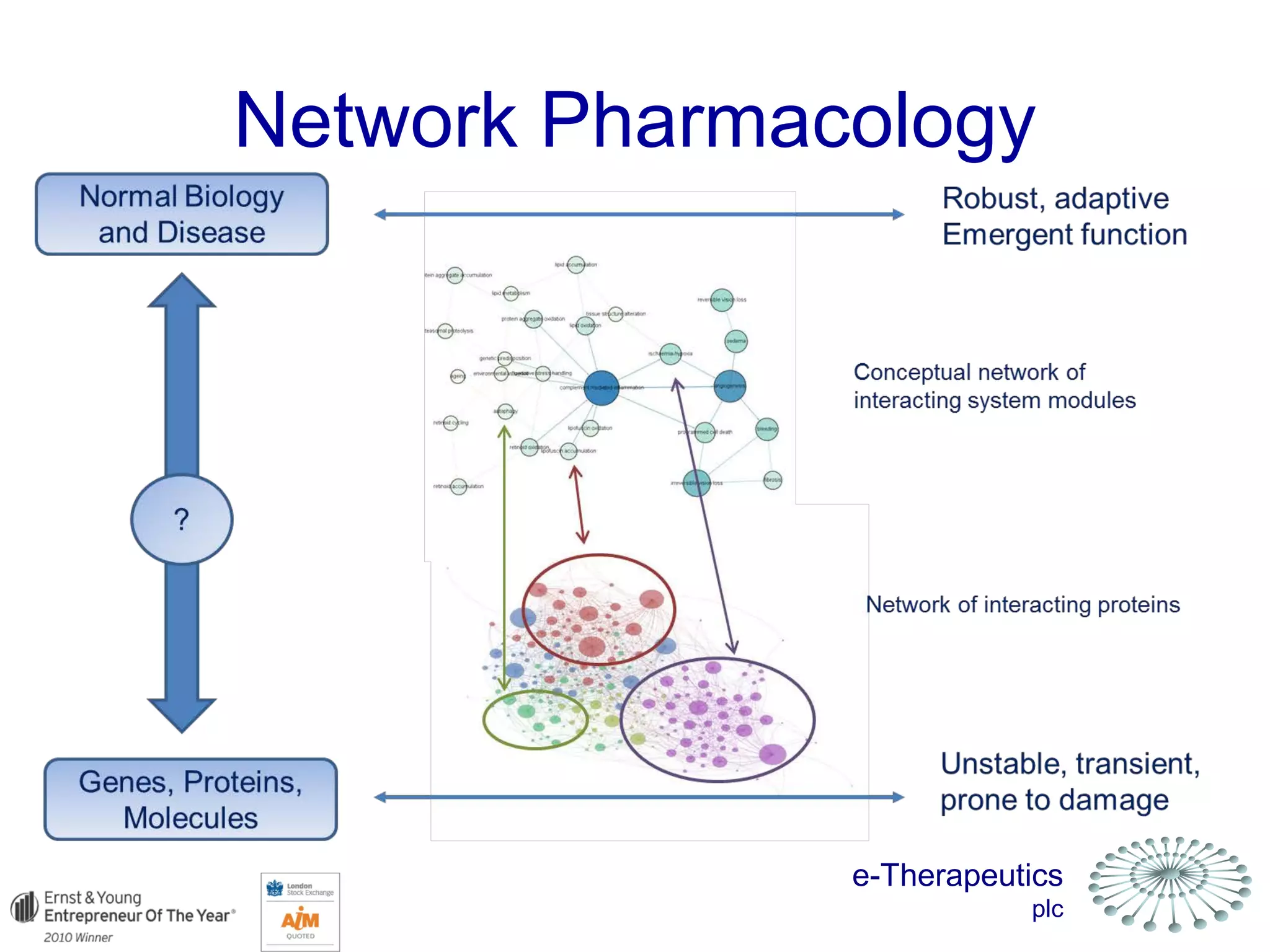
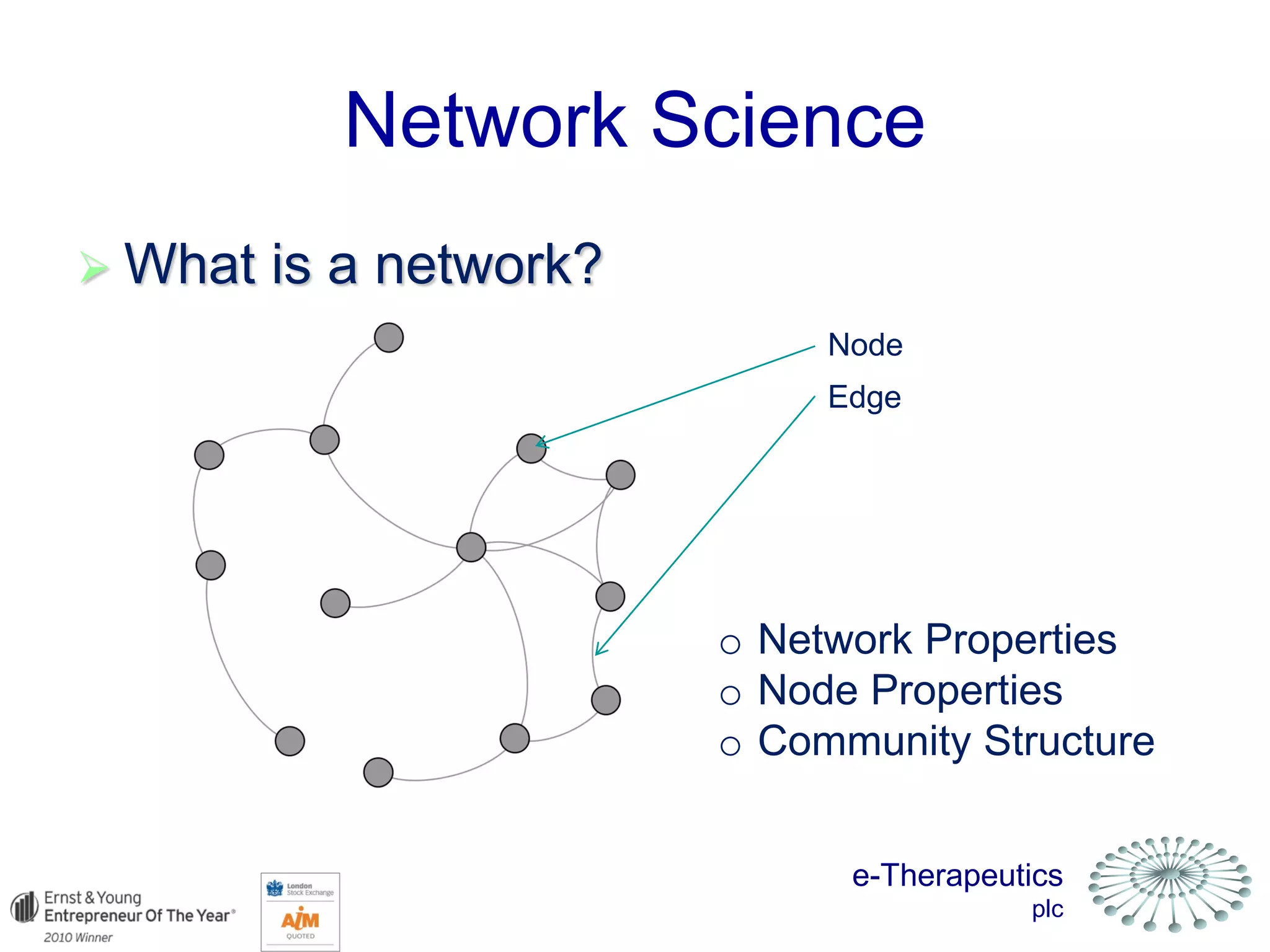
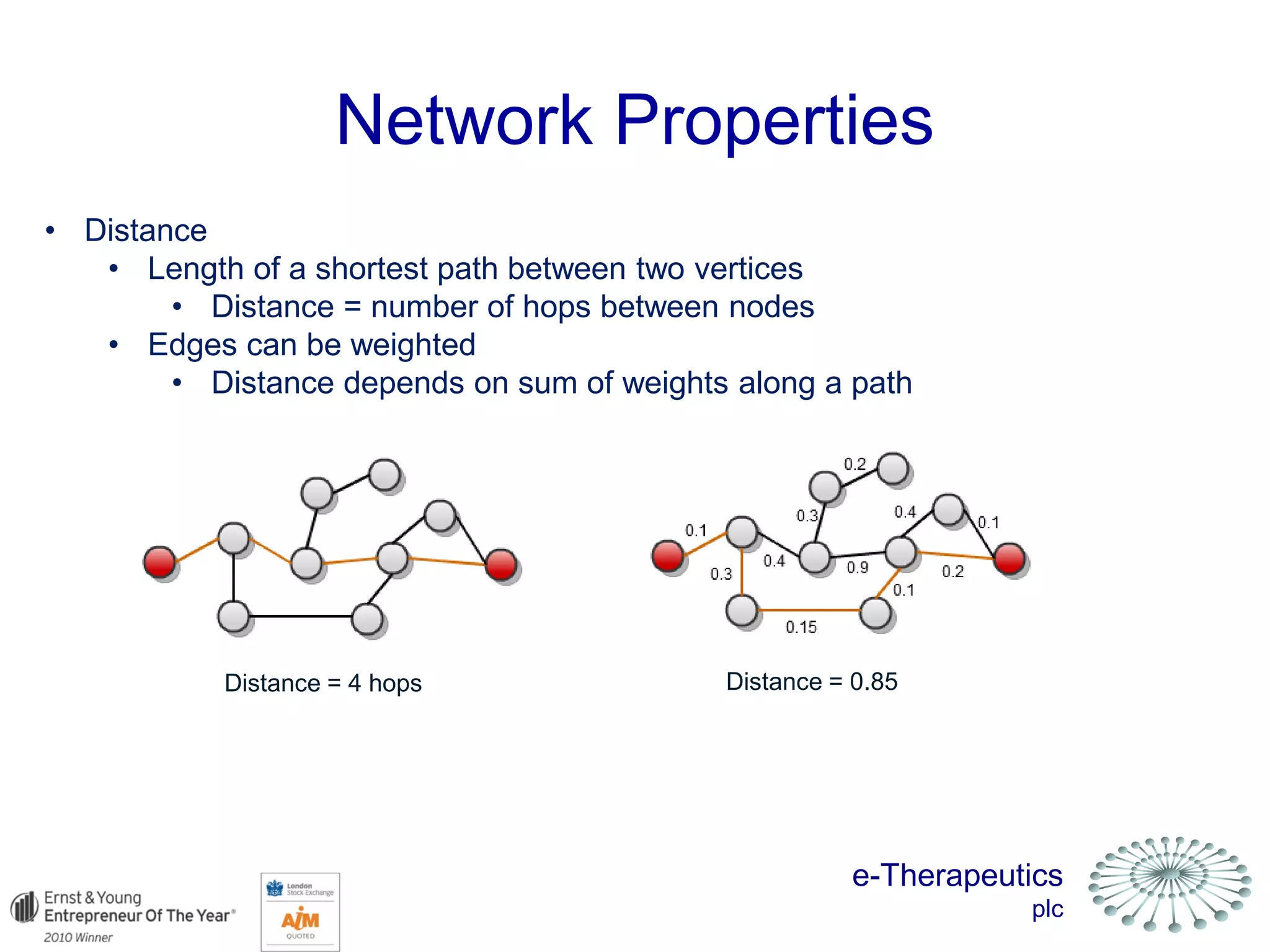
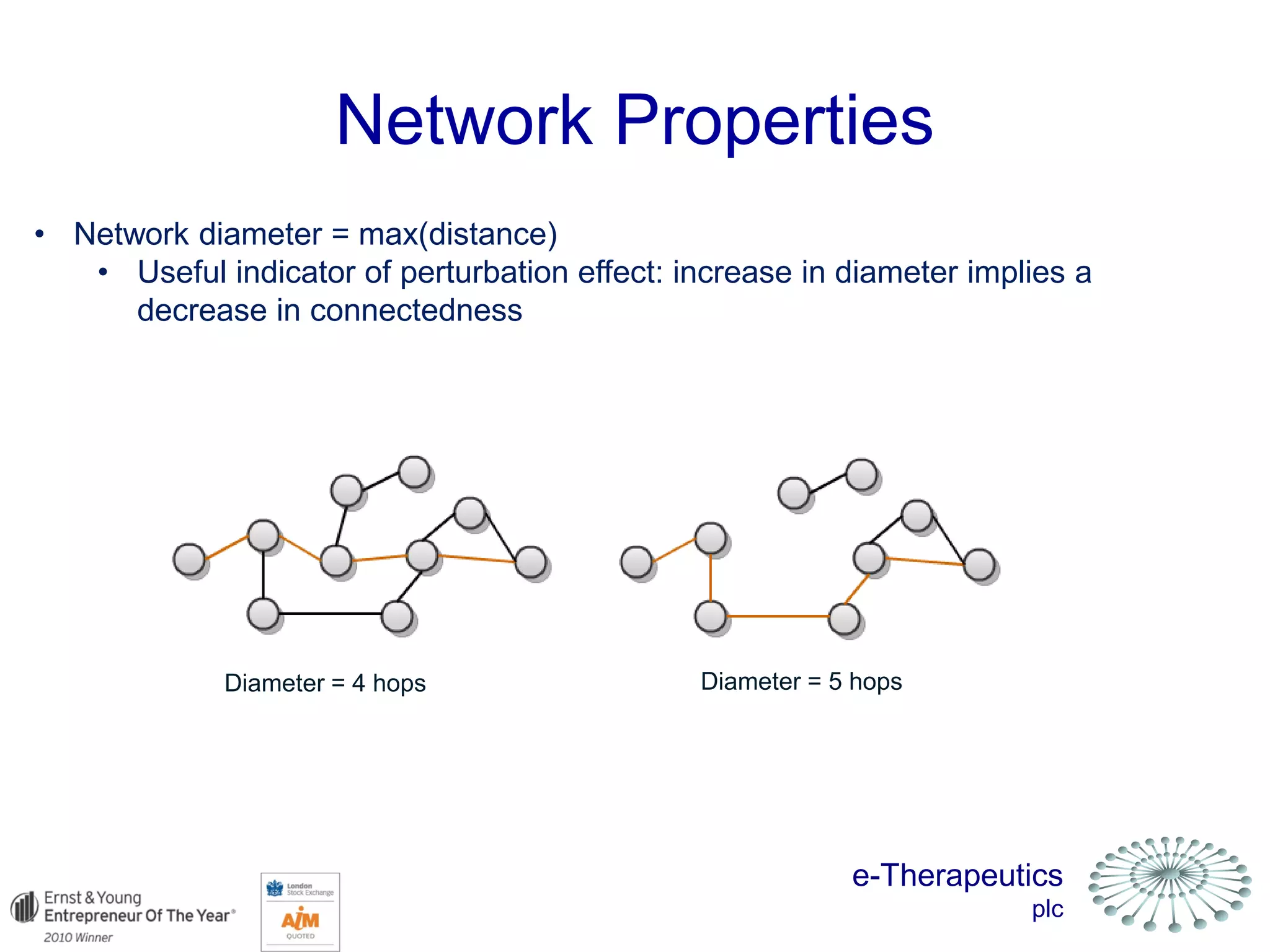
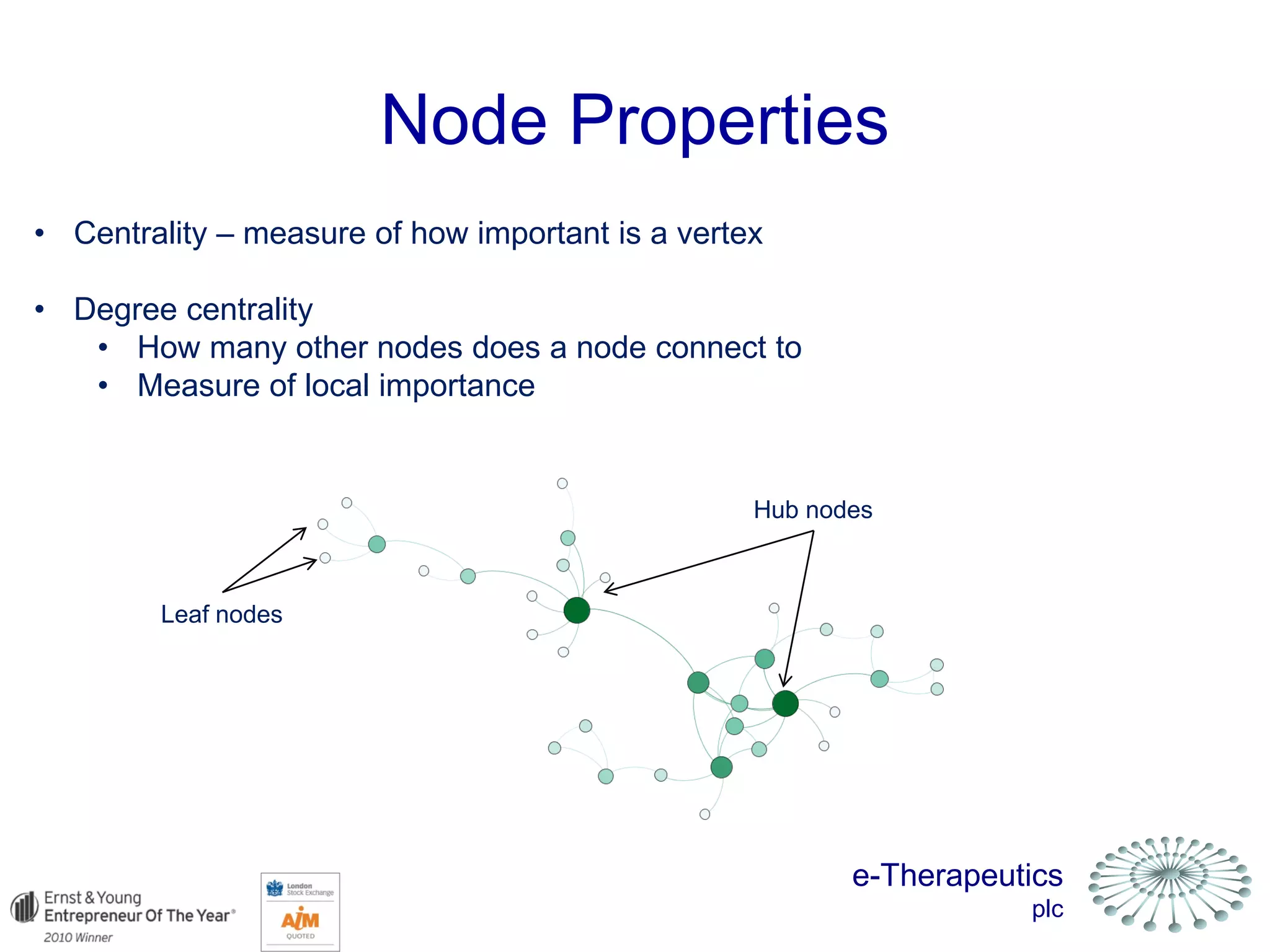
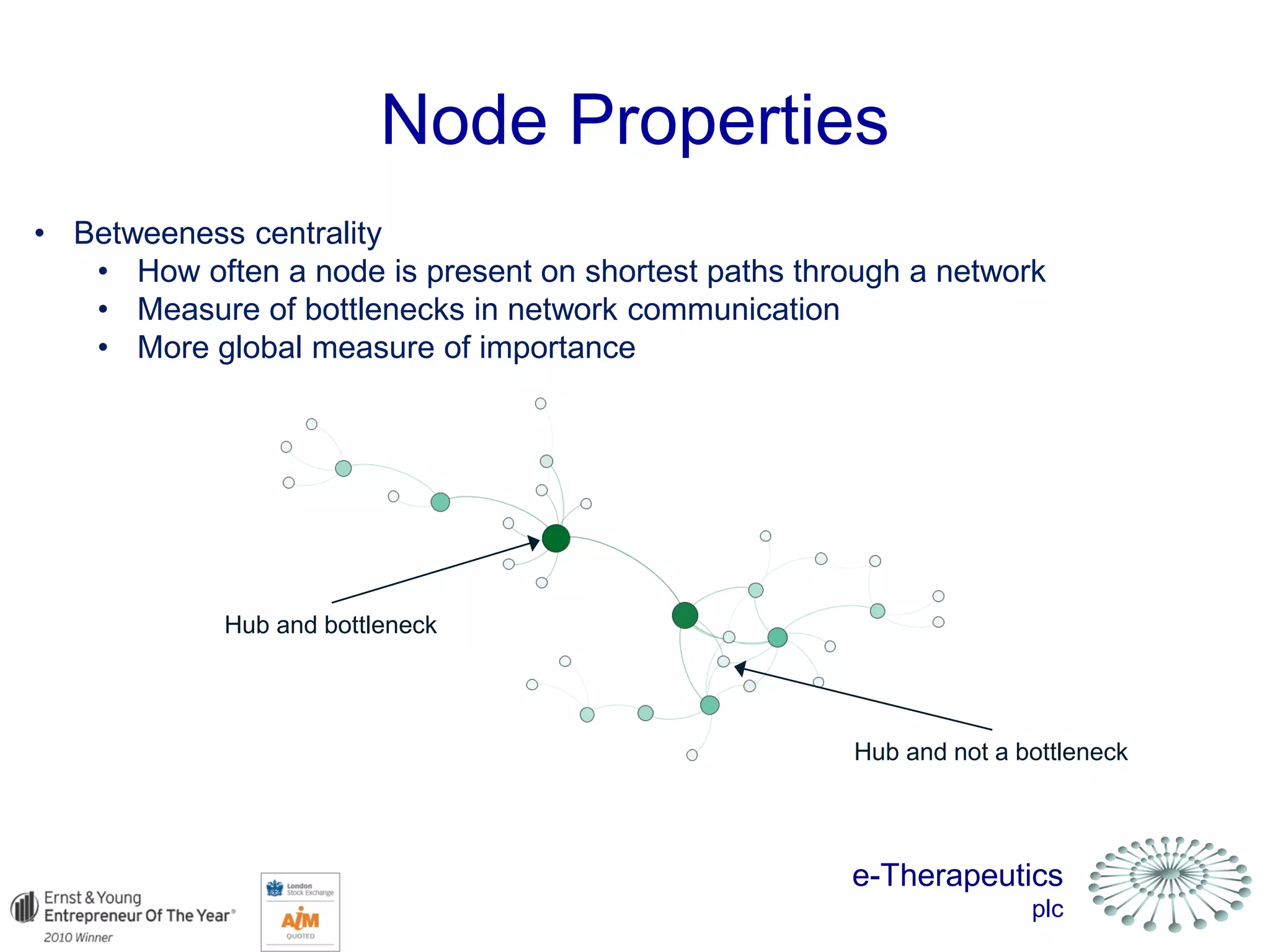
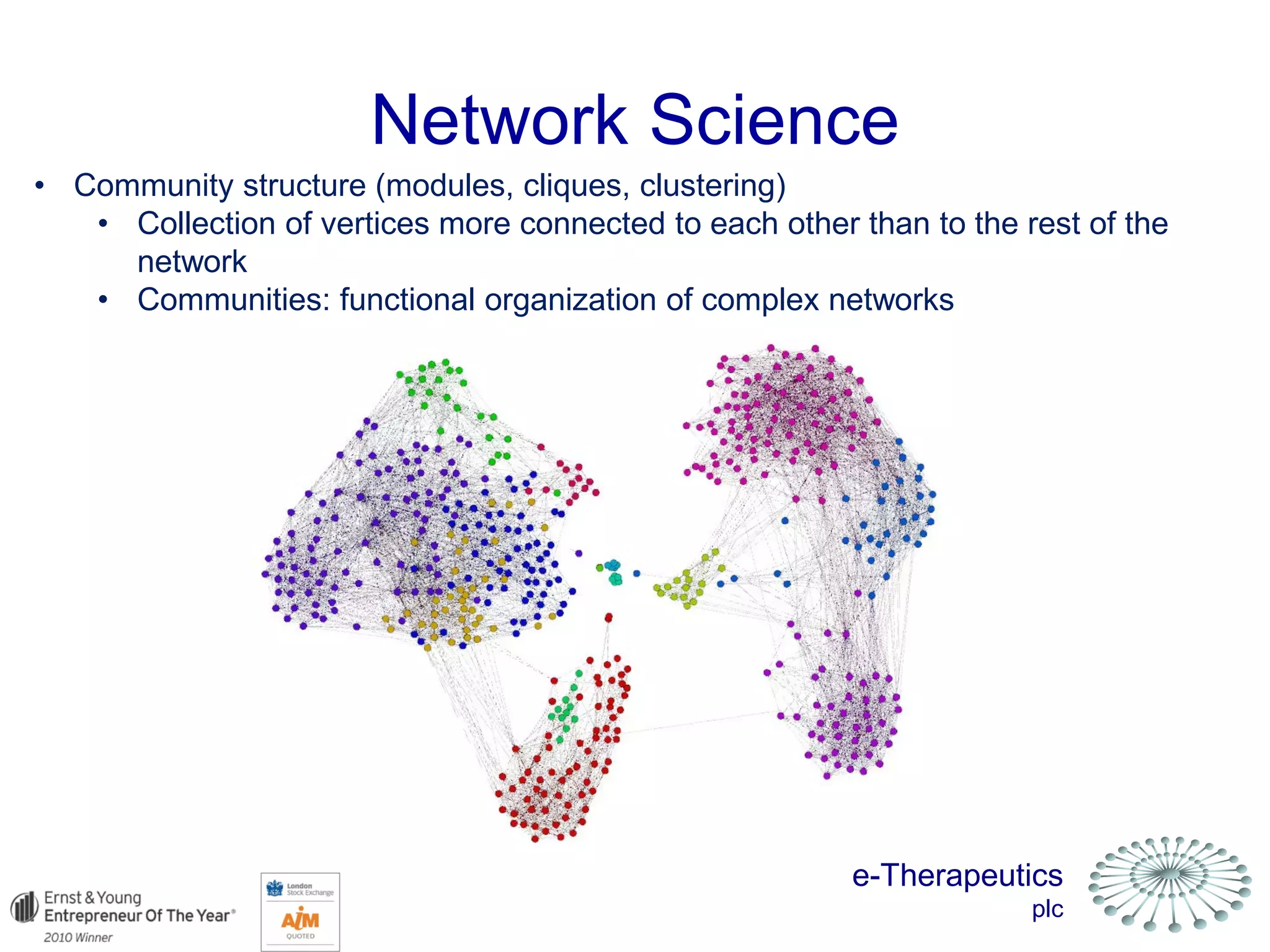
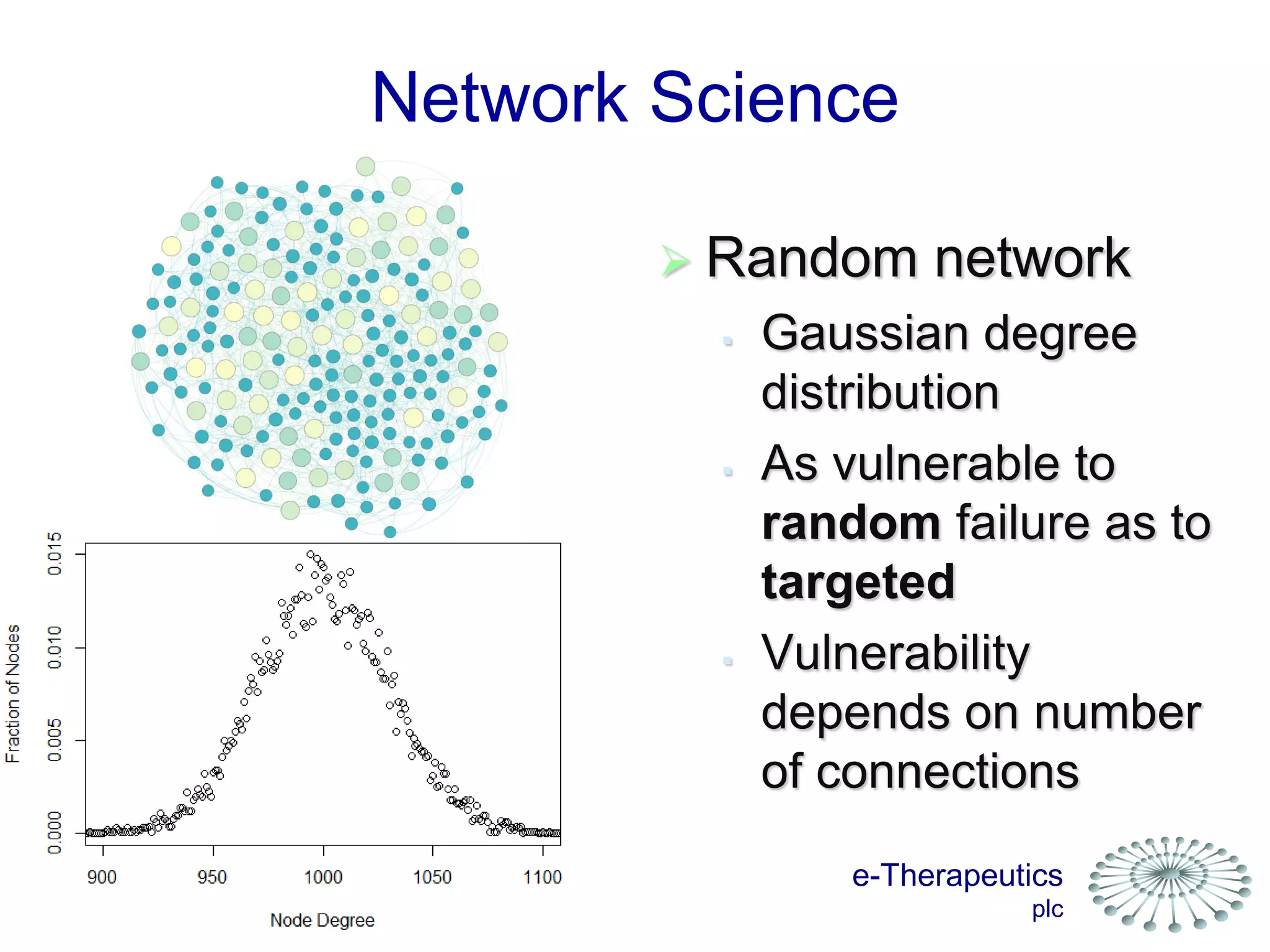
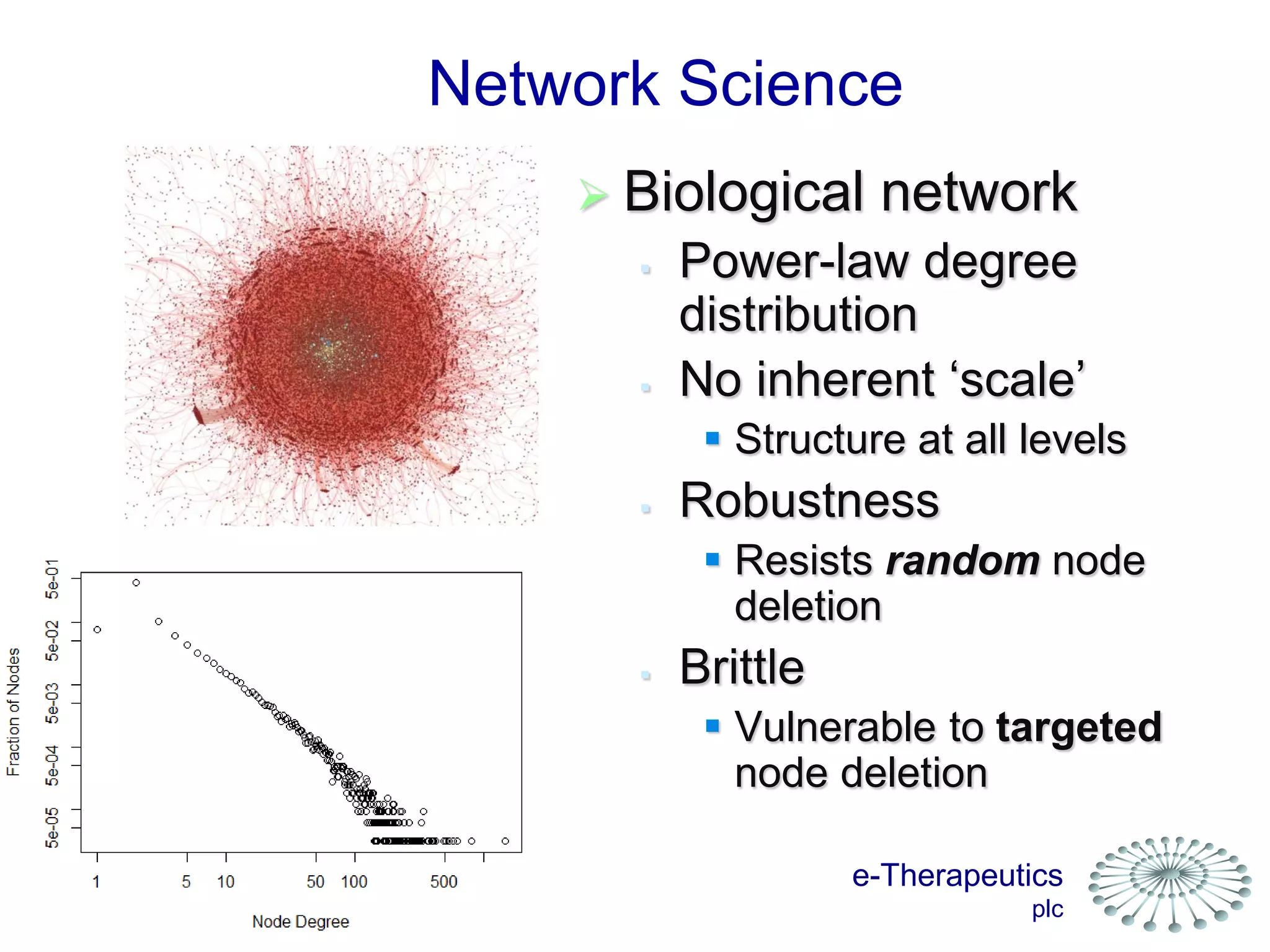
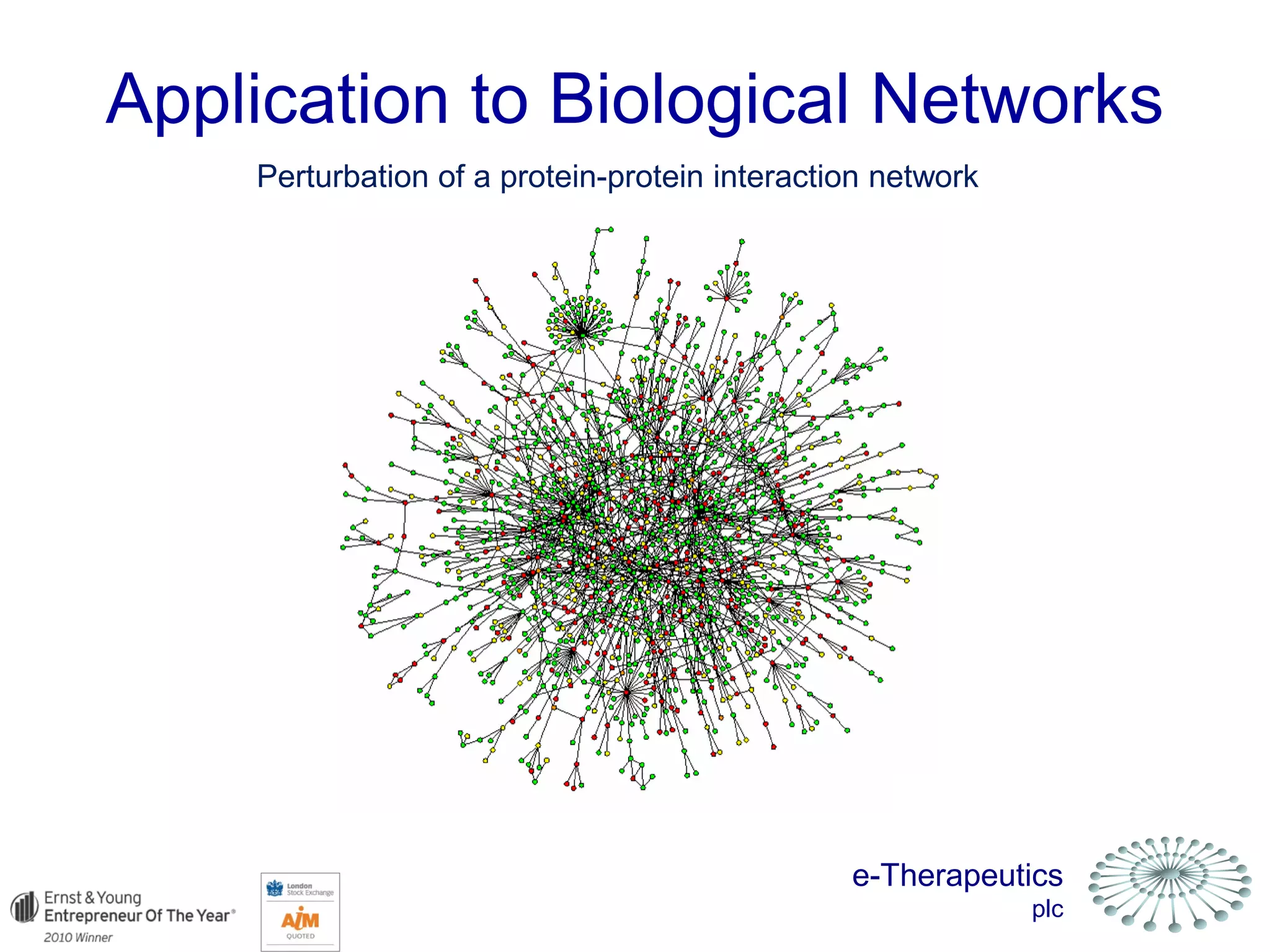
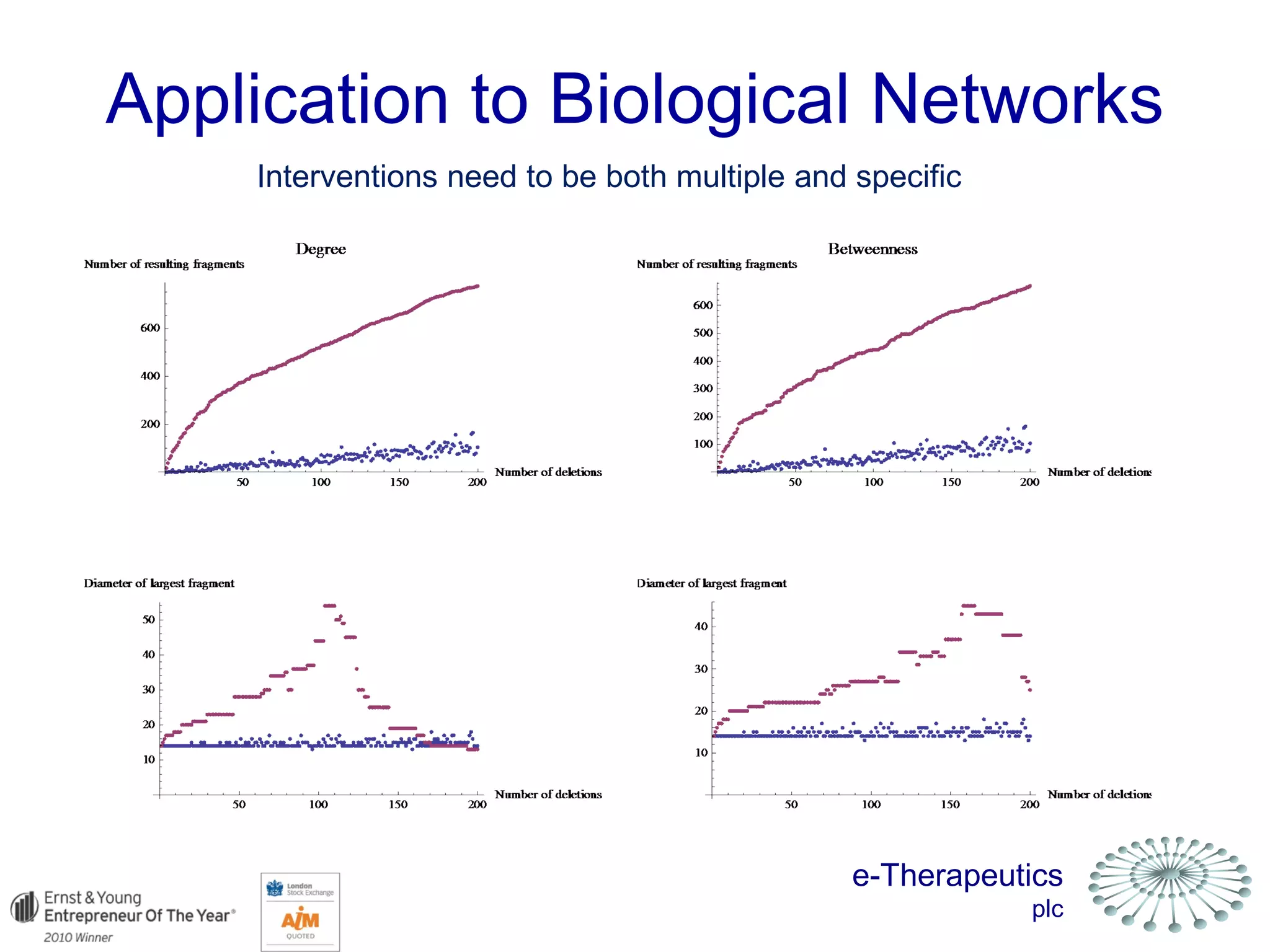
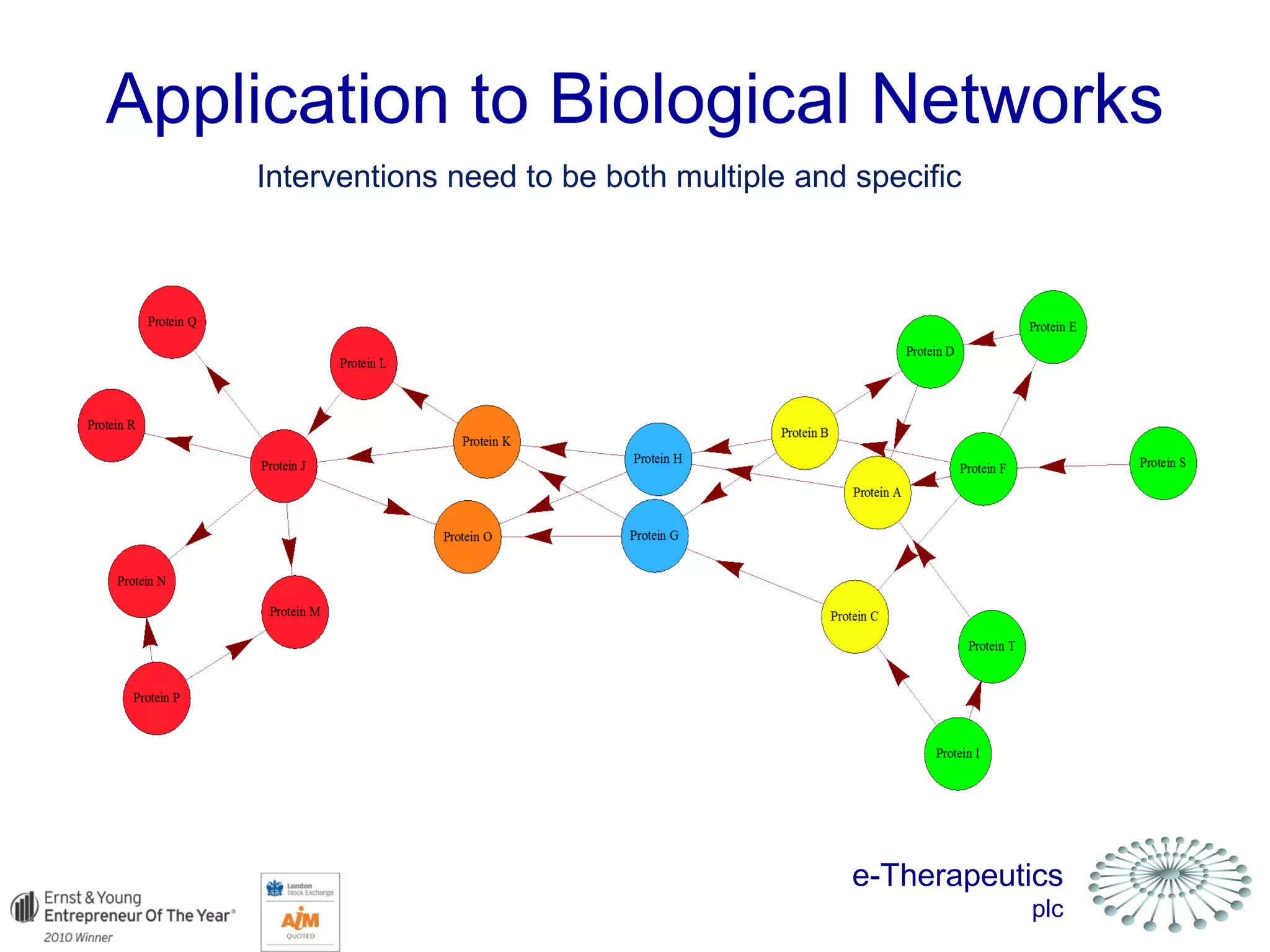
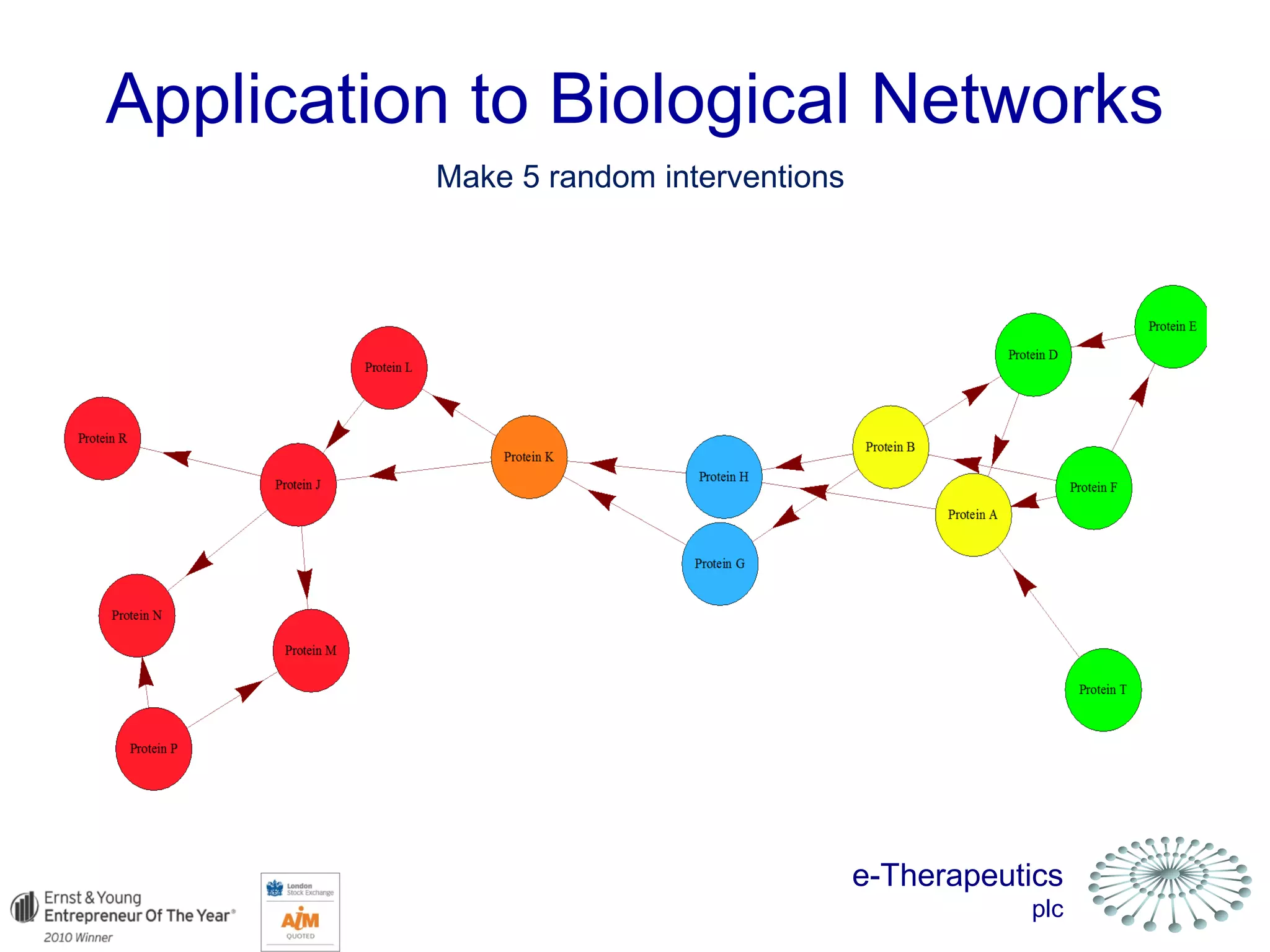
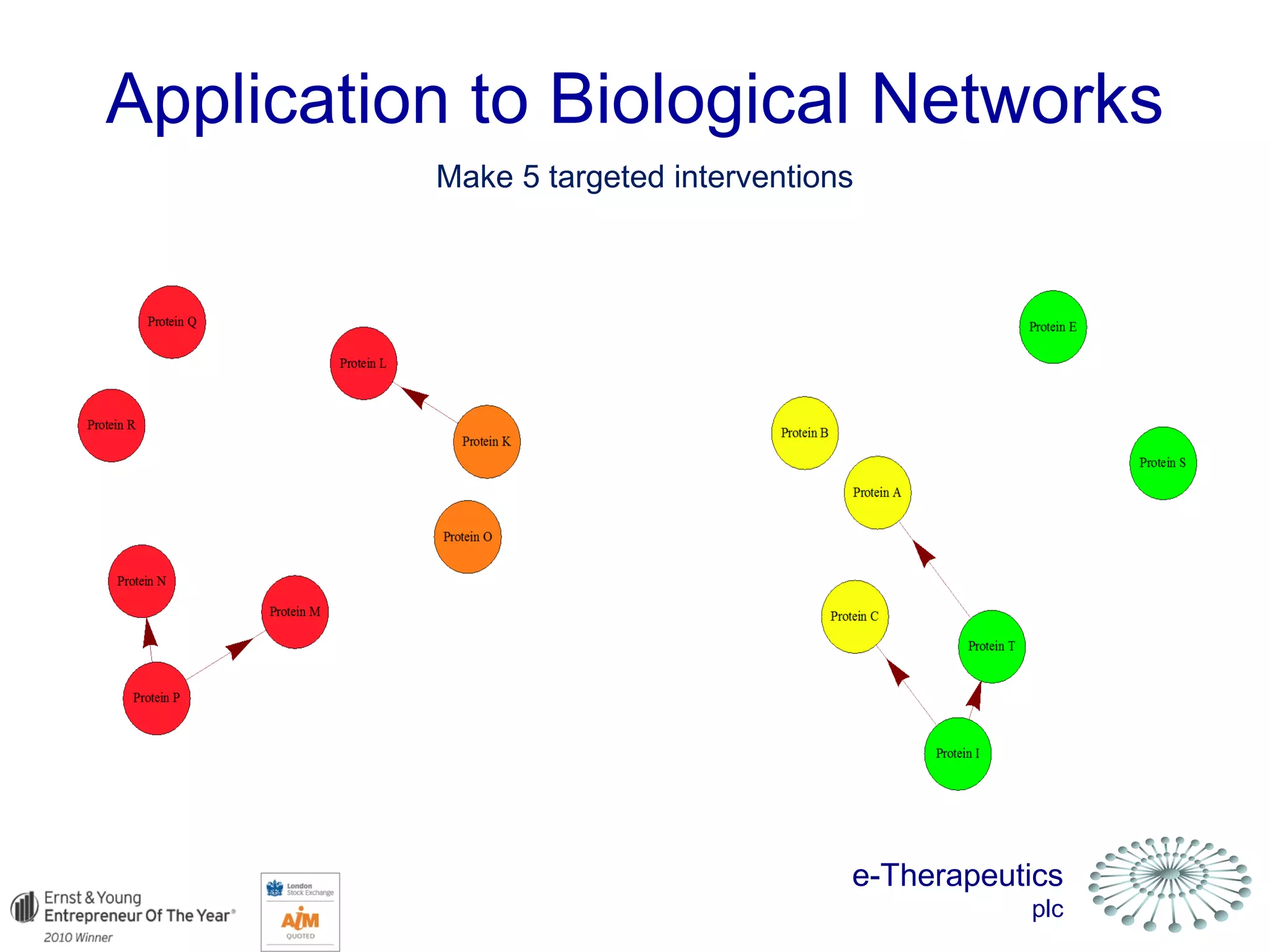
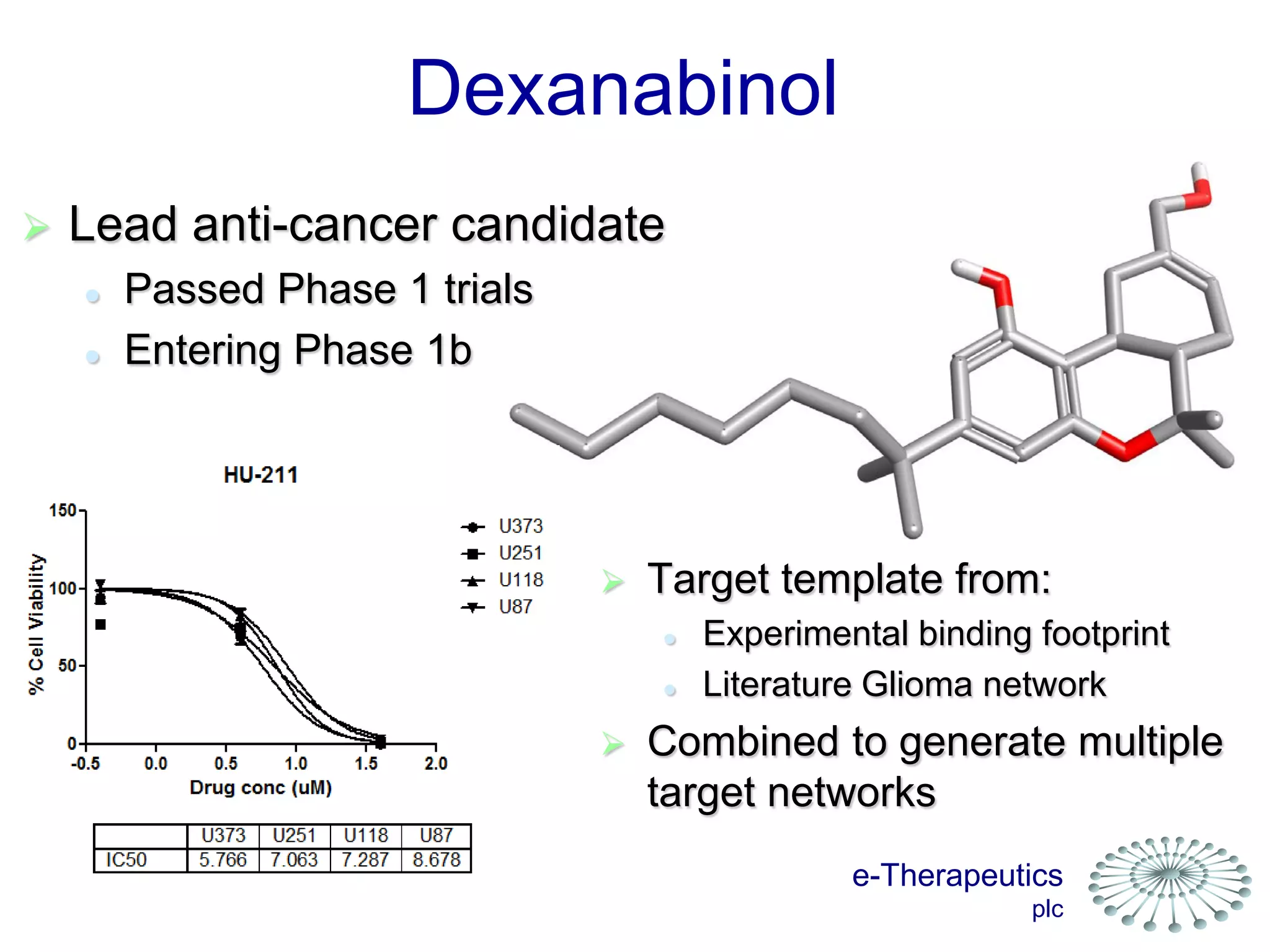
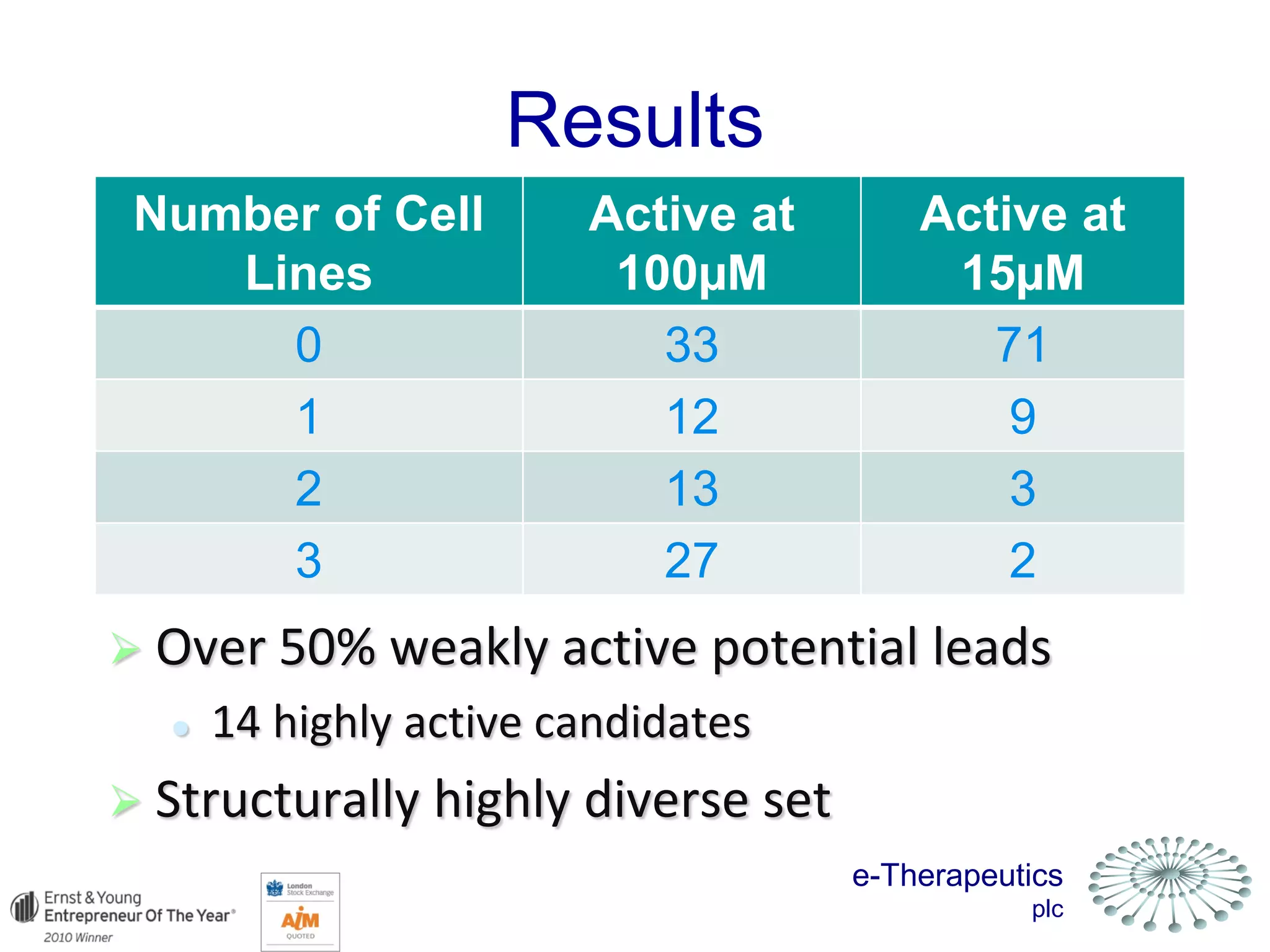
This document discusses using network pharmacology and computational modeling to identify novel potential anti-cancer agents. It describes how E-Therapeutics constructs disease networks and then uses its proprietary chemoinformatics tools to identify compounds that could impact those networks. One lead anti-cancer candidate, Dexanabinol, is highlighted which has passed Phase 1 trials. Experimental validation of compounds predicted to impact glioma networks identified over 50% as weakly active potential leads and 14 as highly active candidates, demonstrating the potential of this network pharmacology approach.