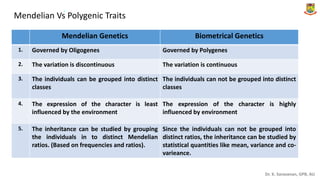
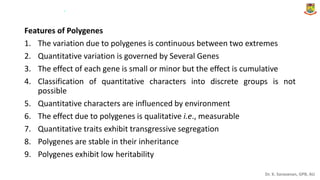
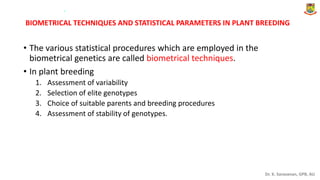
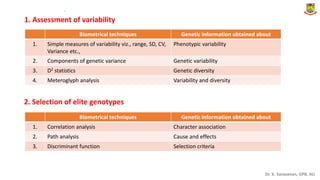
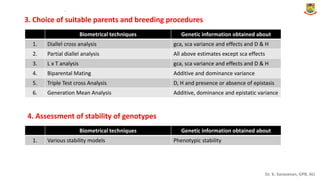
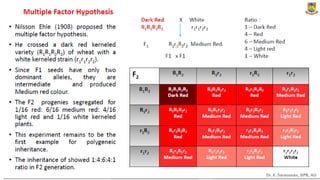
This document provides an introduction to principles of quantitative genetics. It discusses the history and development of the field, beginning with Mendel's foundational work in genetics and Galton's development of statistical techniques. It describes how early geneticists differed in their views of inheritance as qualitative vs quantitative. Key figures who helped establish quantitative genetics are mentioned, including Fisher who integrated Mendelian and statistical approaches. The document outlines differences between Mendelian and polygenic traits. It also discusses types of statistics used in quantitative genetics like first and second degree statistics, as well as biometrical techniques and parameters used in plant breeding like assessment of variability, selection of elite genotypes, choice of parents, and stability analysis.