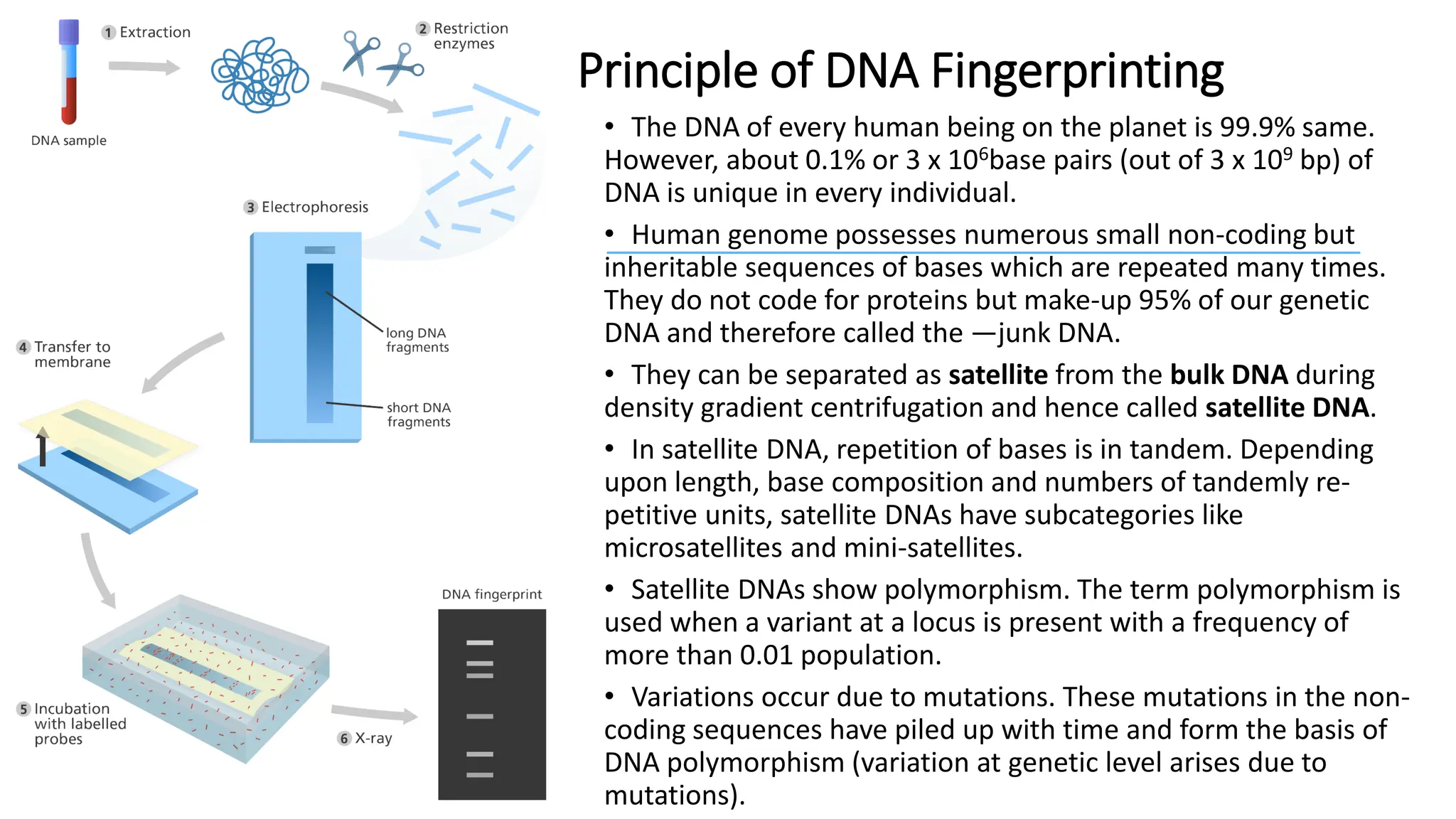
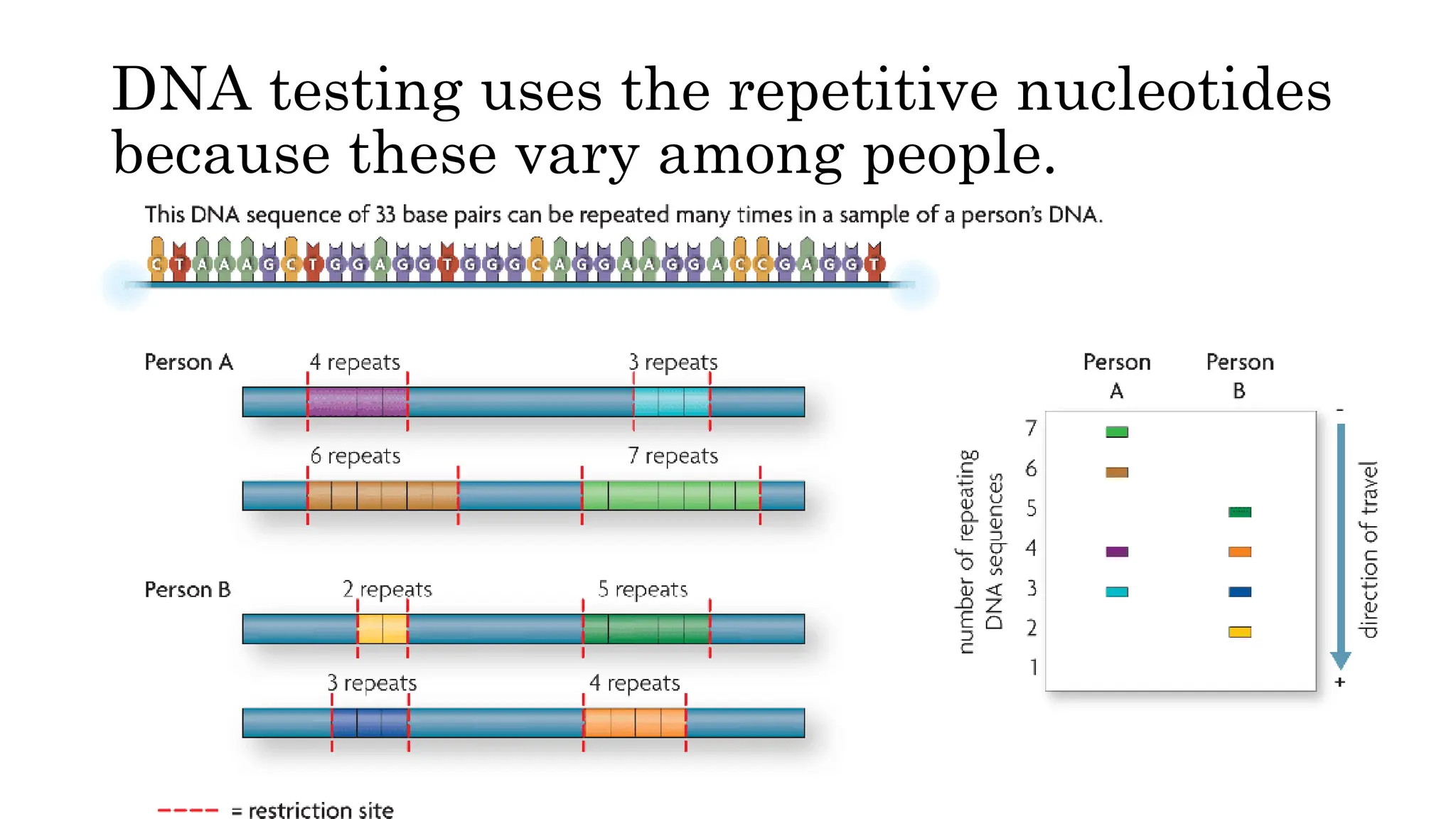
DNA fingerprinting is a technique that identifies living organisms at the molecular level by analyzing unique DNA patterns from individuals. It has applications in criminal investigations and paternity testing, with each person's DNA profile differing due to variations at specific loci caused by mutations. The process involves isolating and replicating DNA, cutting it with restriction enzymes, and separating the resulting fragments using gel electrophoresis.