Embed presentation
Download as PDF, PPTX



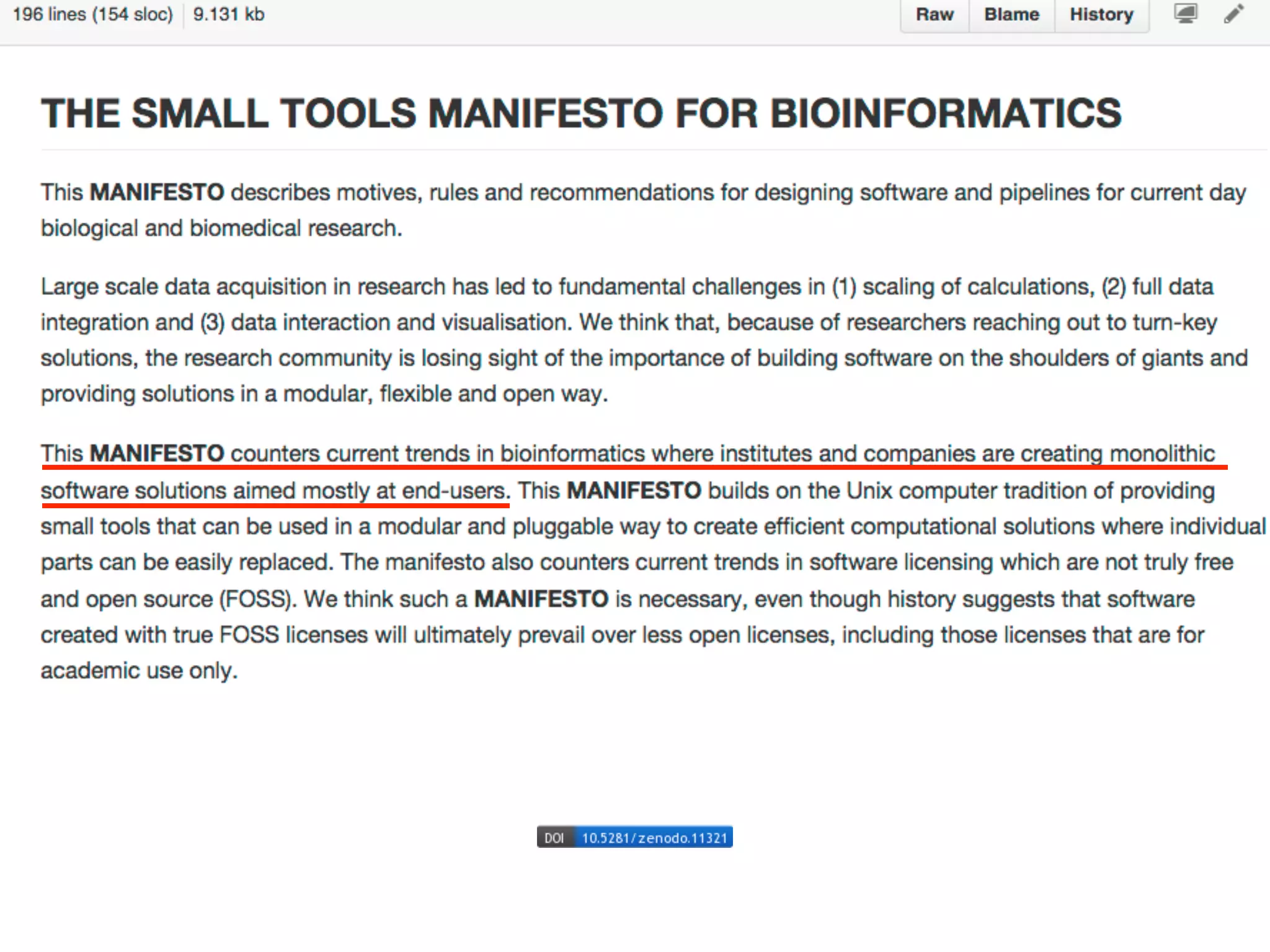









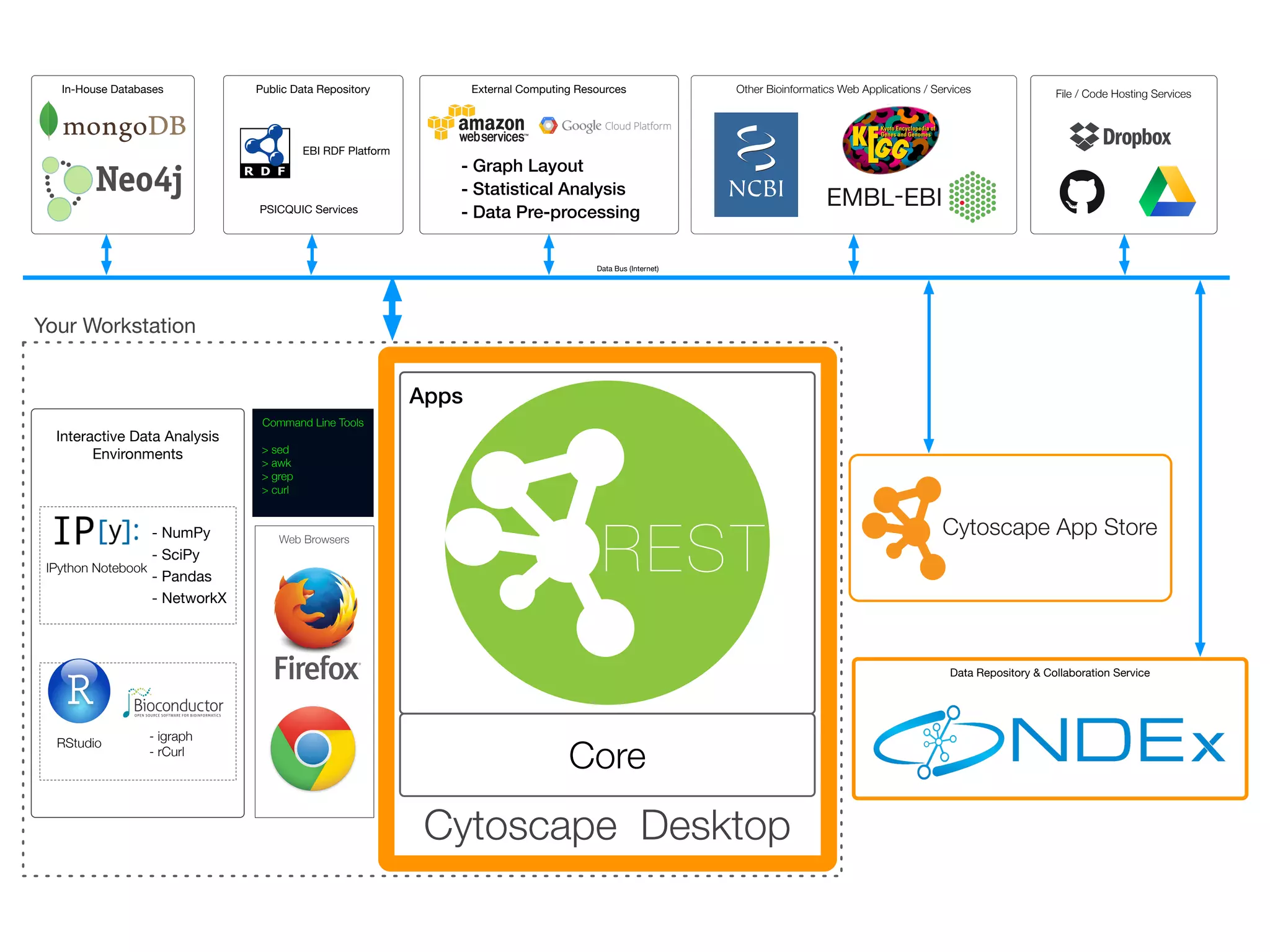




cyREST provides platform-independent access to Cytoscape's data models and functions via REST. This allows different tools like RStudio, IPython notebooks, command line utilities, and web apps to interact with Cytoscape. The goal is for all bioinformatics tools to work seamlessly together. cyREST demonstrates controlling Cytoscape from an IPython notebook to enable interactive data analysis across environments and computing resources.



















