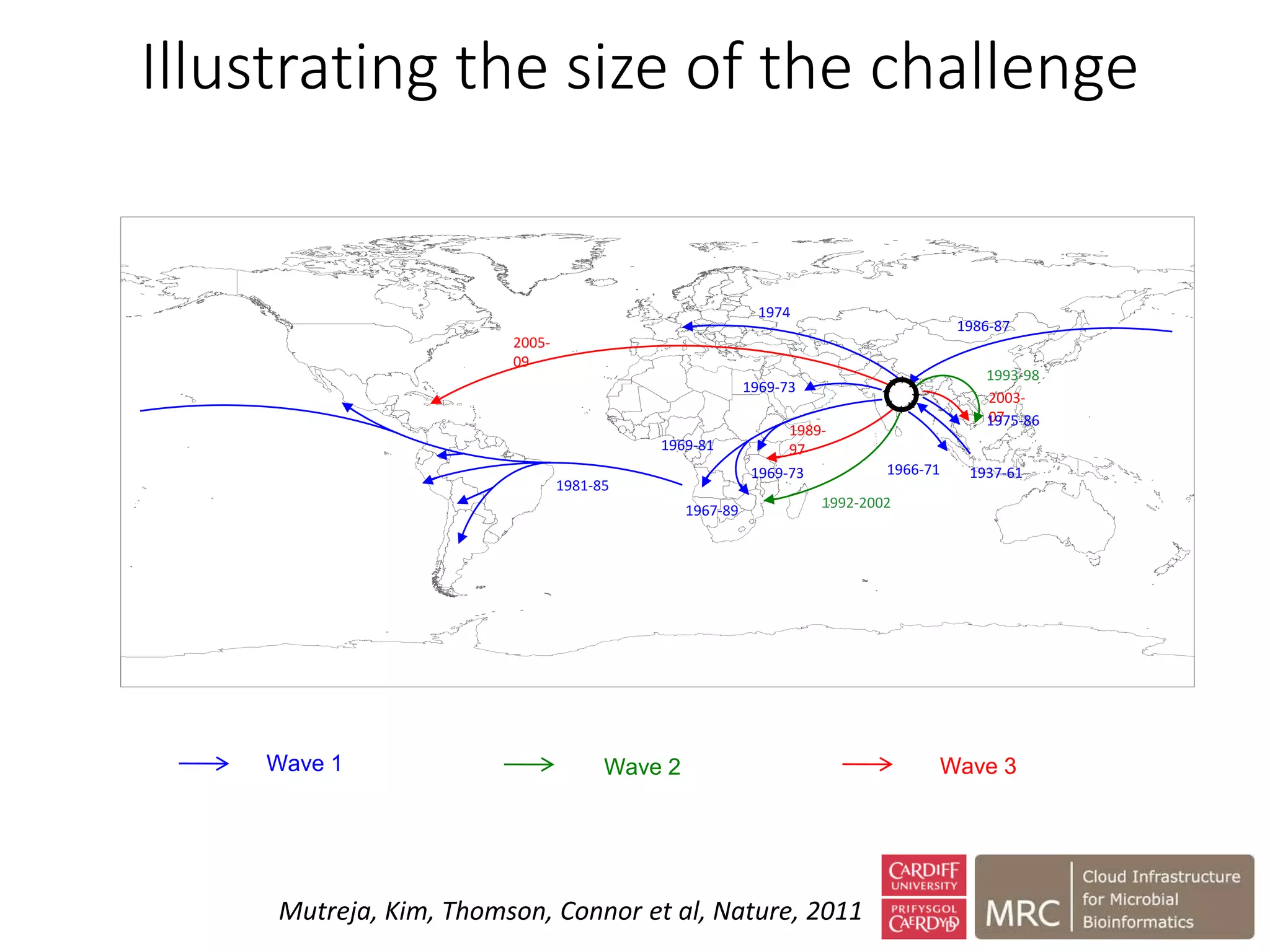
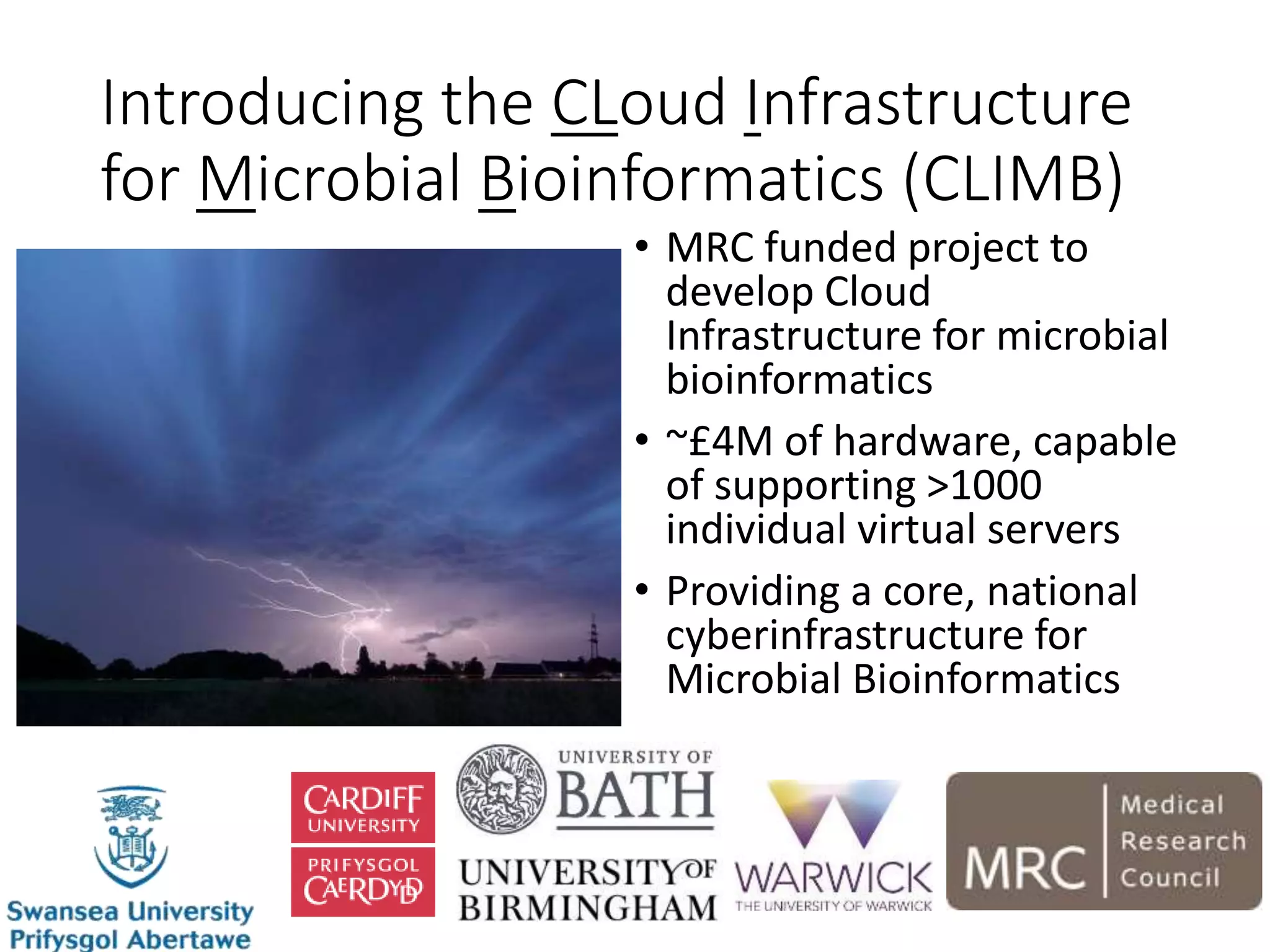
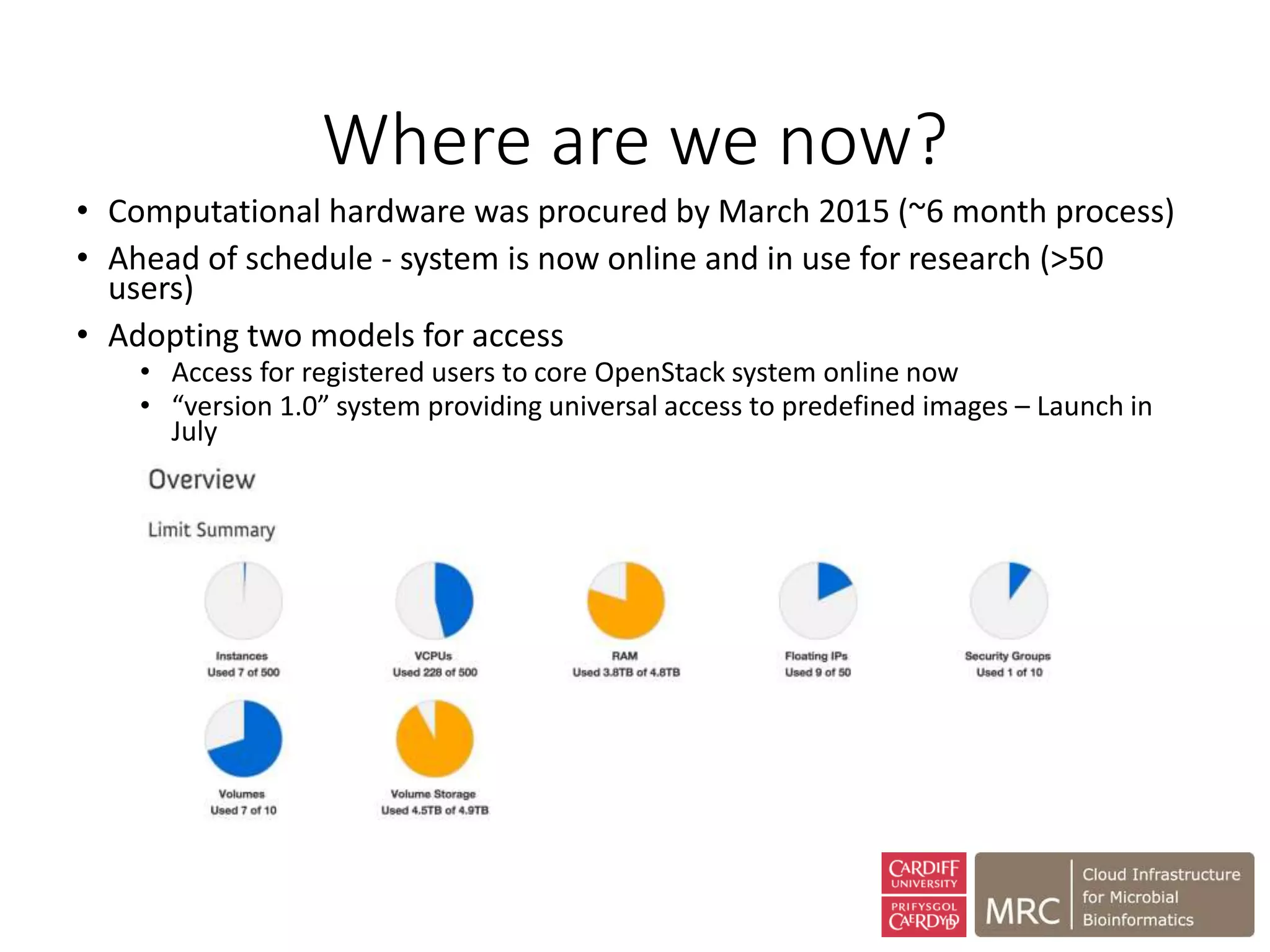
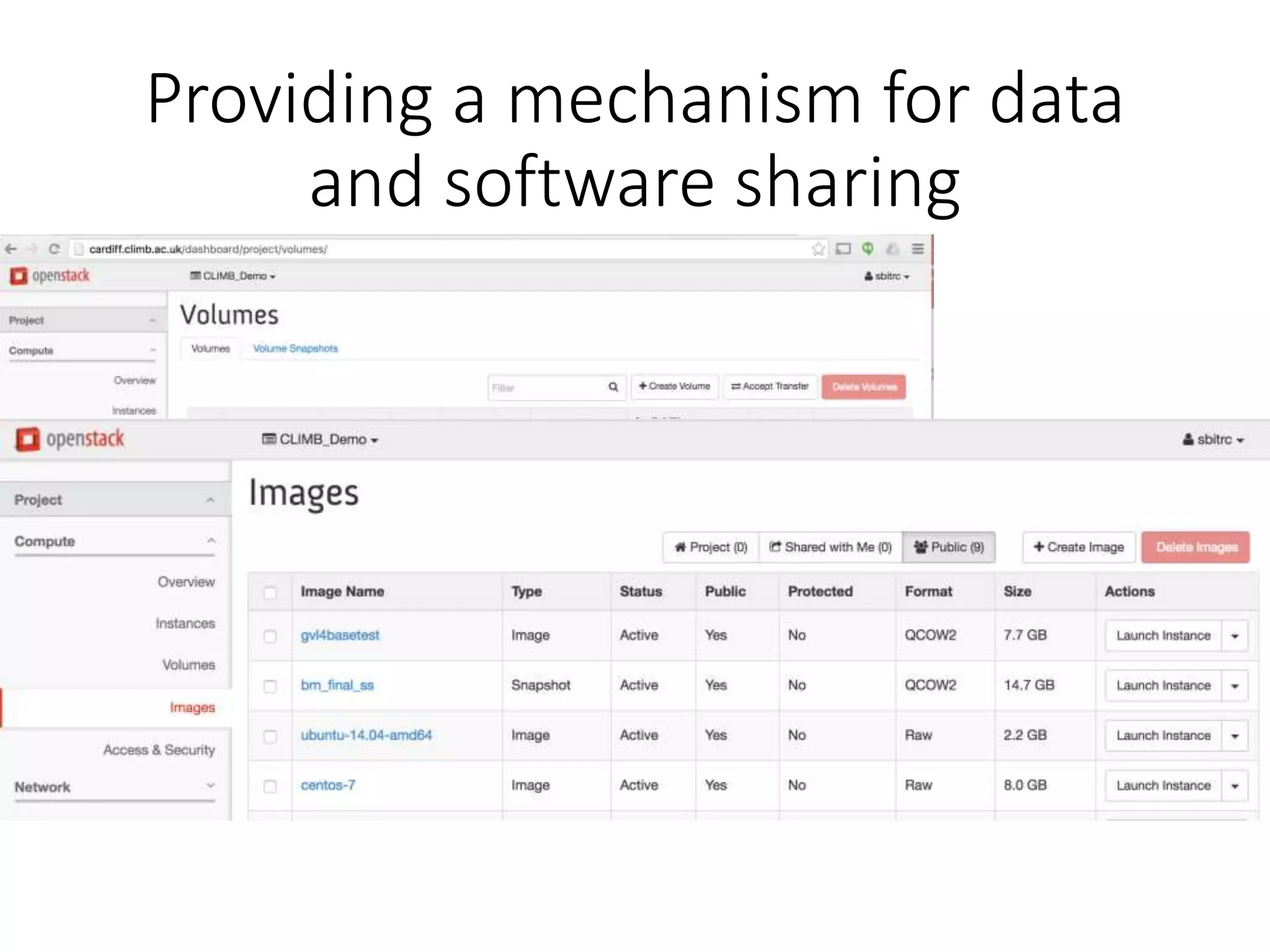
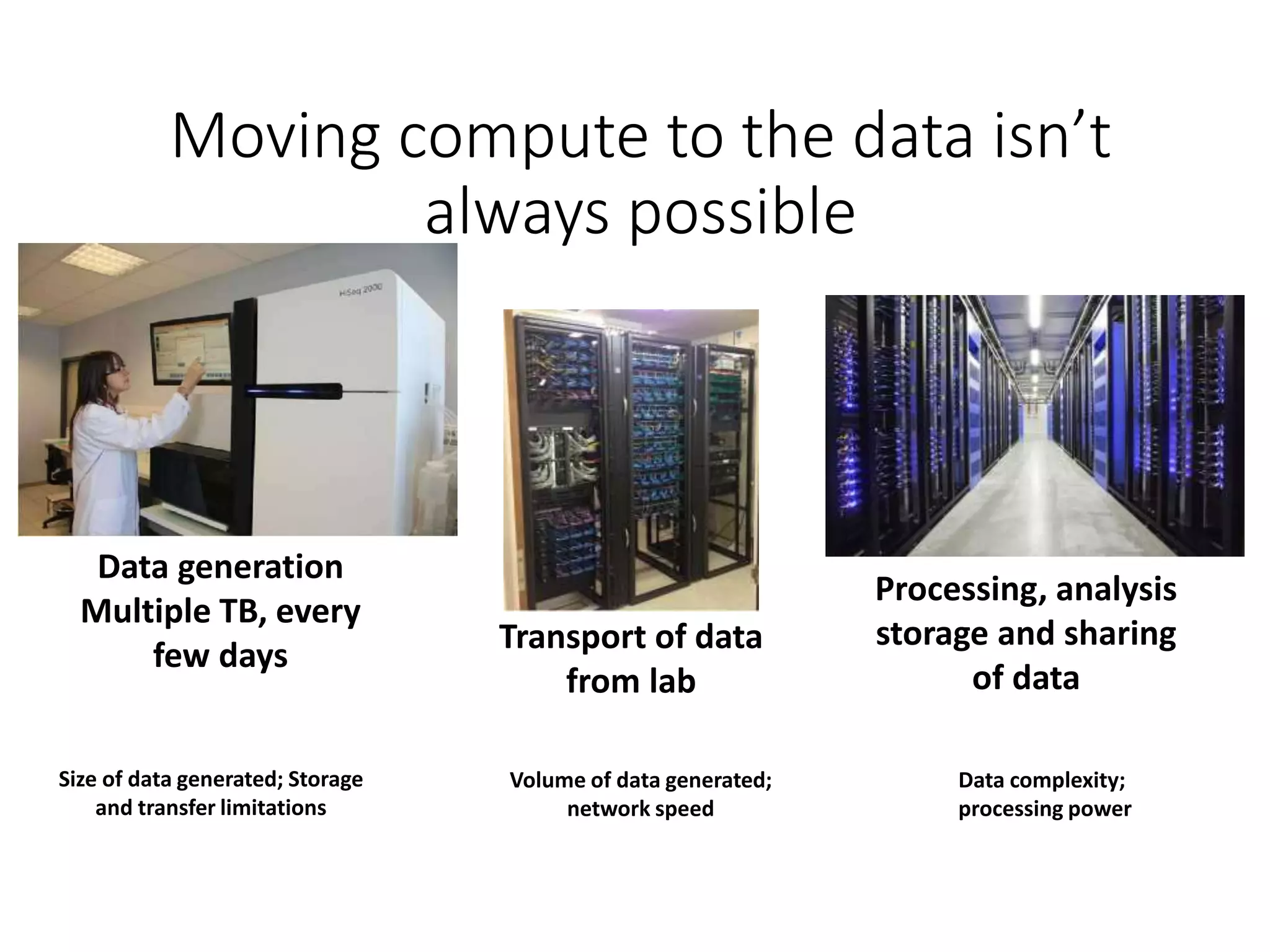
The document outlines the development of a cloud infrastructure for microbial bioinformatics, aimed at improving data sharing and software use across the biology field. It highlights the challenges faced in handling biological big data, such as storage and computational capacity, while emphasizing the importance of cloud-based solutions for facilitating collaboration and resource sharing. The CLIMB project, funded by the MRC, proposes a national cyberinfrastructure capable of supporting over 1,000 virtual servers to enhance microbial research.