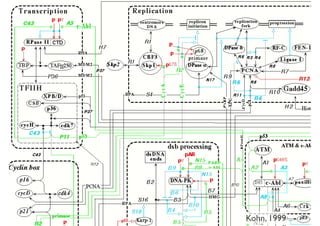
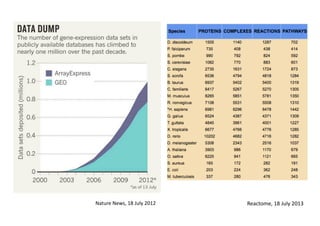
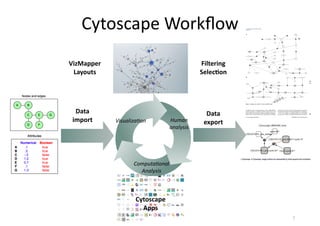
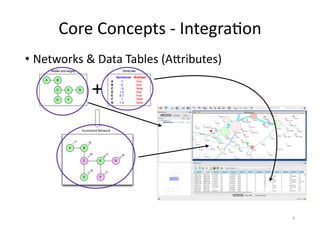
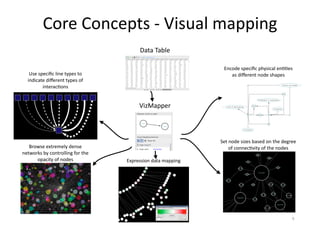
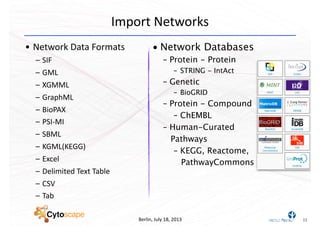
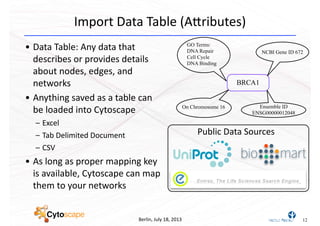
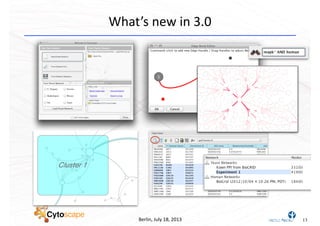
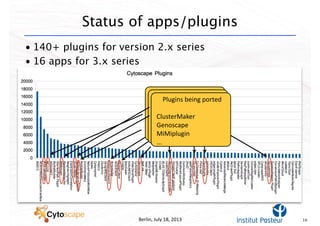
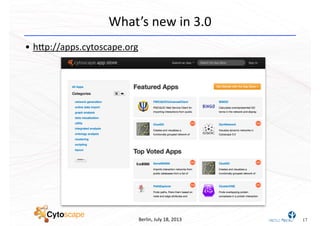
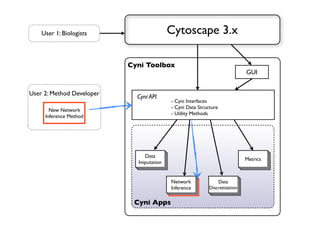
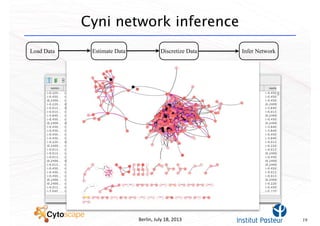
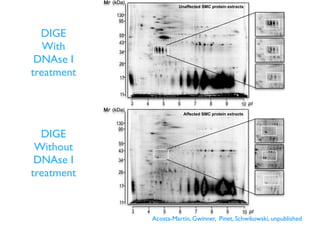
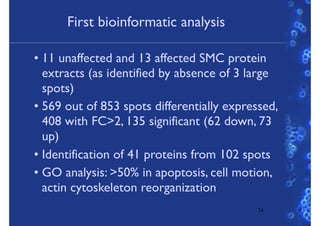
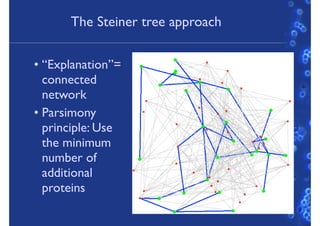
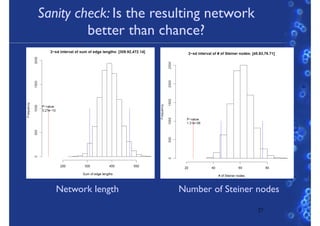
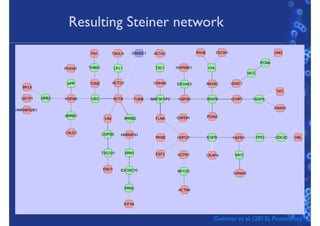
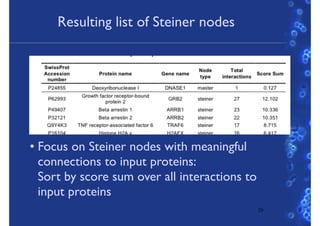
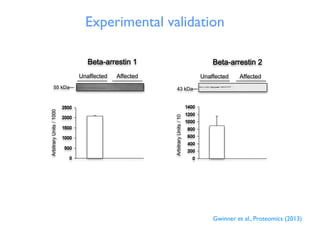
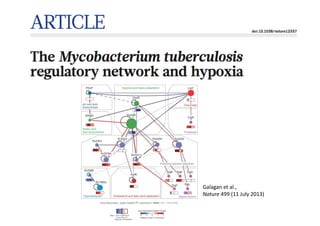
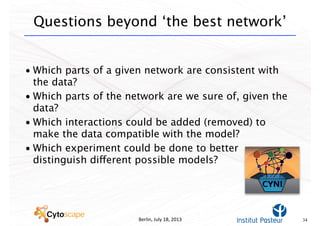
This document discusses computational tools for analyzing biological networks and molecules. It begins by describing how networks can be inferred from molecular data using the Cytoscape platform and its apps. It then discusses how networks can help identify proteins of interest by describing a case study where network analysis identified additional proteins that provided an "explanation" for differences observed in proteomic data. The document concludes by discussing how computational analysis can help address questions that go beyond simply identifying the best network, such as which parts of a network are well-supported or could be modified to better fit the data.