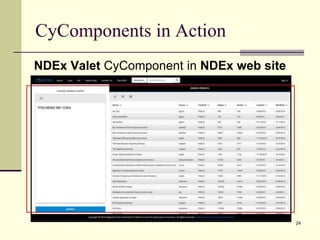
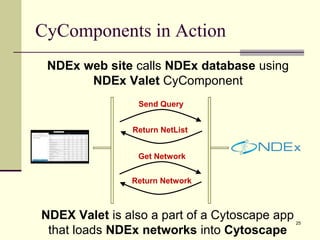
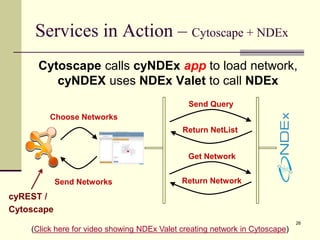
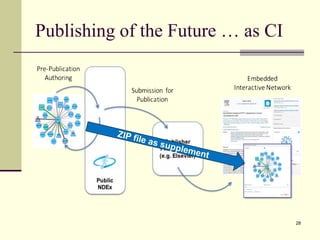
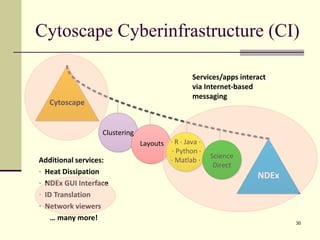
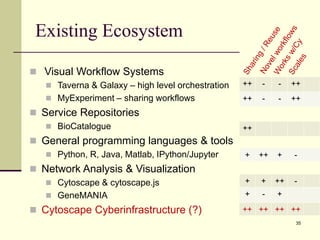
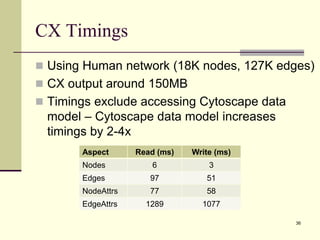
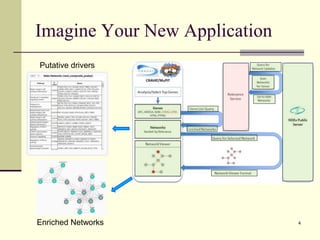
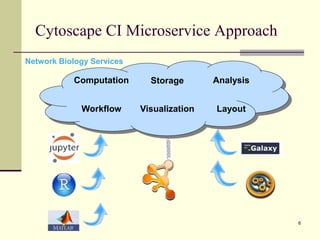
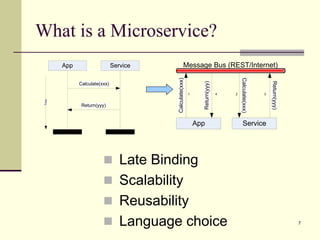
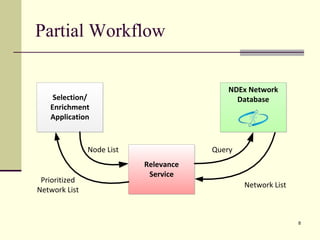
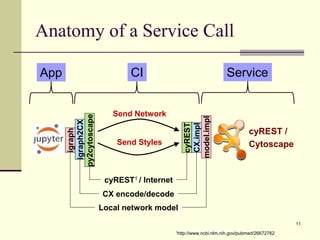
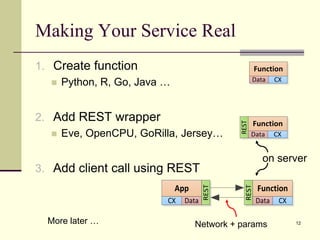
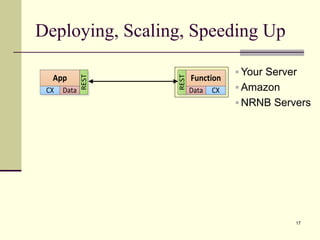
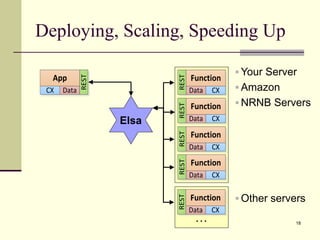
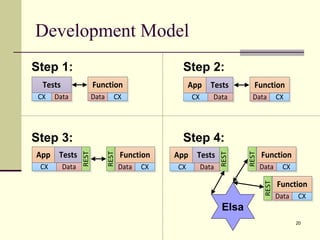
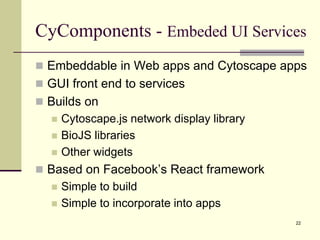
The document outlines the challenges and solutions in developing network biology software, emphasizing the need for reusable components and novel workflows to improve speed and efficiency. It introduces a microservice-based architecture within Cytoscape for rapid and scalable network analysis, enabling the integration of various programming languages and tools. Additionally, it highlights the importance of community involvement and the sharing of resources to enhance the development and application of network biology services.






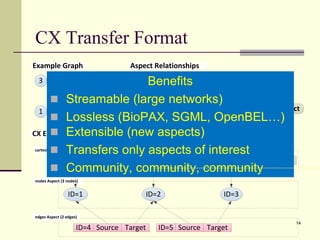
![A CX Snippet
[
{"nodes": [{"@id": "_:1"}, {"@id": "_:2"}]},
{"edges": [{"source": "_:1", "@id": "_:4", "target": "_:2"}]},
{"cartesianLayout": [{"x": "100", "node": "_:1", "y": "100"}]},
{"cartesianLayout": [{"x": "200", "node": "_:2", "y": "300"}]},
{"nodes": [{"@id": "_:3"}]},
{"edges": [{"source": "_:2", "@id": "_:5", "target": "_:3"}]},
{"cartesianLayout": [{"x": "100", "node": "_:3", "y": "200"}]}
]
15](https://image.slidesharecdn.com/cytoscapecichapter2-160815140646/85/Cytoscape-CI-Chapter-2-15-320.jpg)



![Reusing CyComponents is Easy
23
Can be used from Cytoscape app or Web app
function init(loginInfo) {
const cyto = CyFramework.config([NDExStore]);
cyto.render(NDExLogin, document.getElementById('valet'), {
defaults: loginInfo,
onSubmit: () => {
const state = cyto.getStore('ndex').server.toJS();
connect(state);}
});
}](https://image.slidesharecdn.com/cytoscapecichapter2-160815140646/85/Cytoscape-CI-Chapter-2-23-320.jpg)