Embed presentation
Download as PDF, PPTX



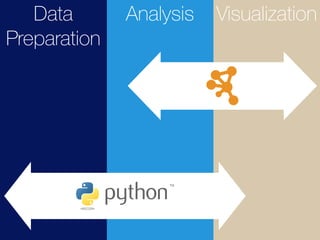
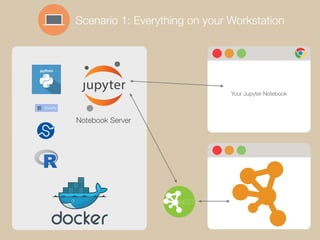
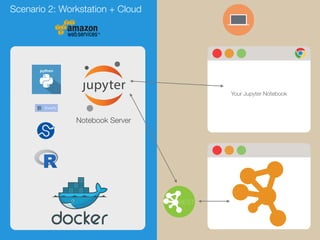




The document discusses building reproducible network data analysis and visualization workflows using REST APIs and containerization. It aims to solve problems with complex software stacks that are difficult to set up and not reproducible. The goal is to create reproducible and scalable "dry experiments" using Docker containers, GitHub for source code sharing, Jupyter notebooks as electronic lab notebooks, and the cyREST module for the Cytoscape network analysis software. Examples of scenarios using local workstations and cloud computing are presented, as well as a demo and future plans.










