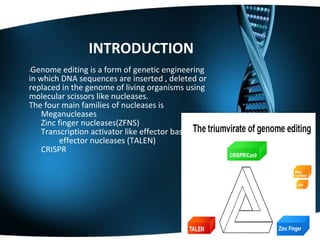
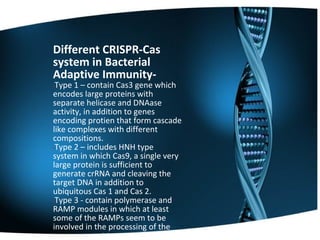
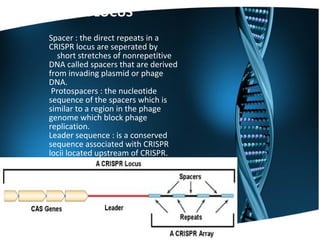
The document summarizes key aspects of CRISPR genome editing technology. It describes how CRISPR uses Cas9 and guide RNA to precisely target and edit DNA sequences. It provides a brief history of CRISPR discovery and outlines its components and mechanism of action. The document also discusses several medical applications of CRISPR including treating Duchenne muscular dystrophy, beta-thalassemia, and testing for viruses. It concludes that CRISPR is a flexible and accurate gene editing tool being explored for various applications in agriculture, biotechnology, and medicine.