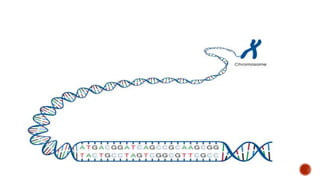
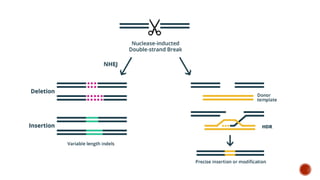
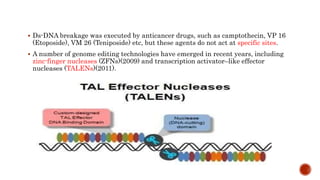
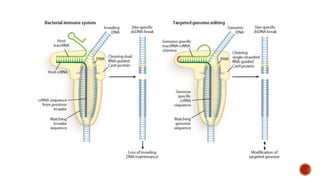
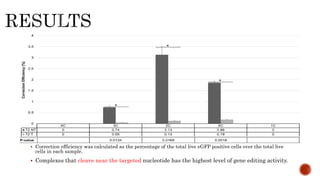
Gene editing uses tools like CRISPR to precisely alter DNA sequences. CRISPR uses guide RNA to target specific DNA sites for editing. Early gene editing tools included ZFNs and TALENs but CRISPR is easier to implement. While CRISPR shows promise for curing diseases, it also raises ethical concerns about altering human heredity that require open discussion.