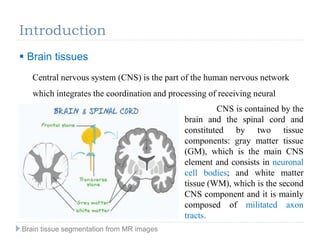
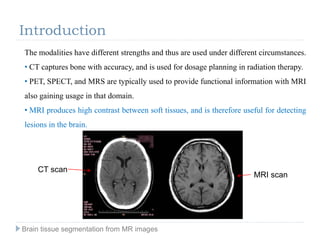
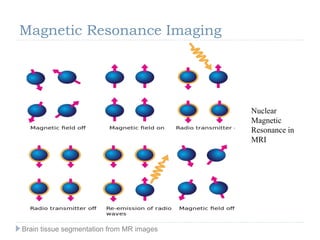
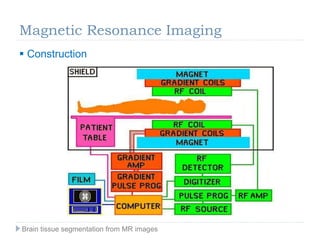
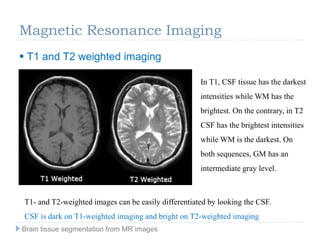
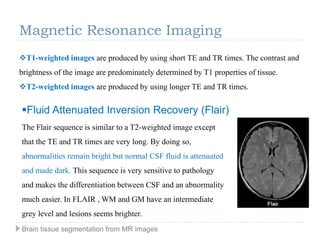
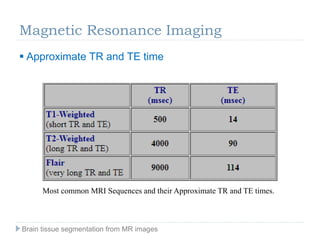
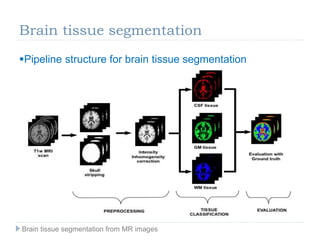
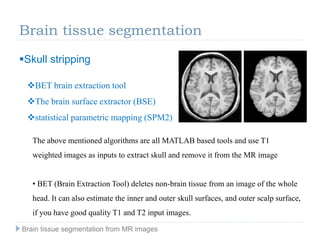
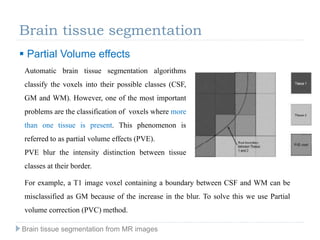
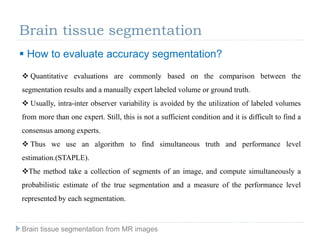
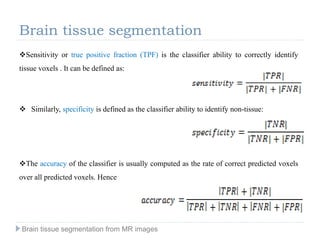
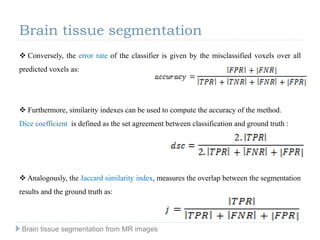
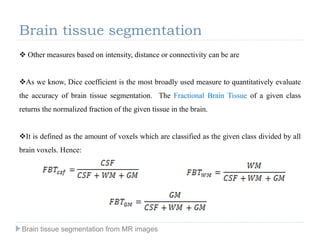
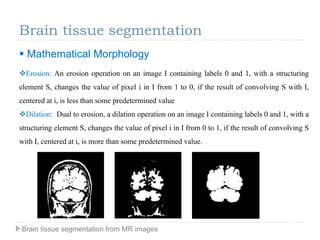
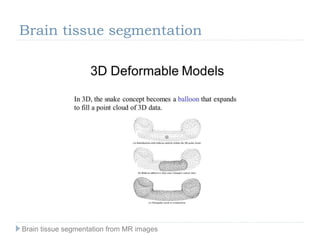
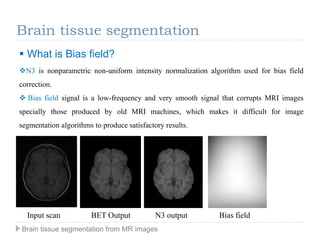
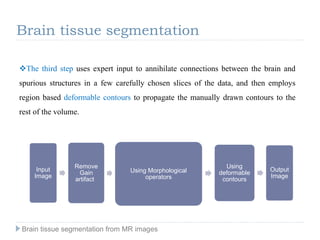
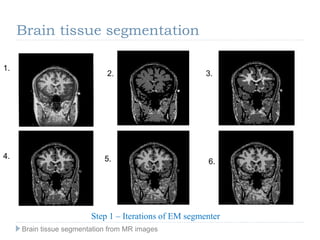
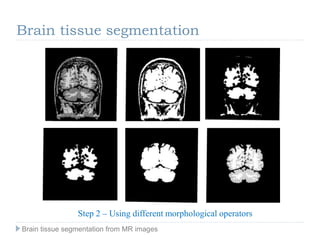
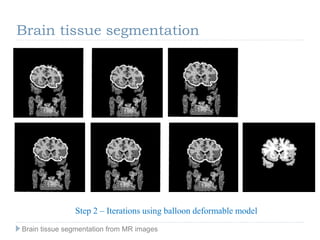
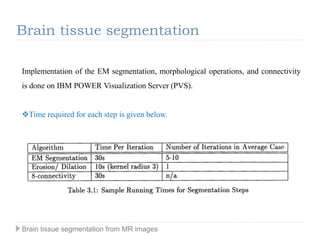
The document discusses the challenges and methodologies of brain tissue segmentation from MRI images, highlighting the need for automated processes due to the complexity of brain structures. It reviews various imaging modalities and their applications, and details the segmentation pipeline, including skull stripping, intensity inhomogeneity correction, and the evaluation of segmentation accuracy. Key techniques mentioned include binary morphology, deformable models, and expectation maximization for classification of different tissue types.