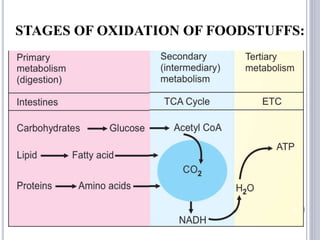
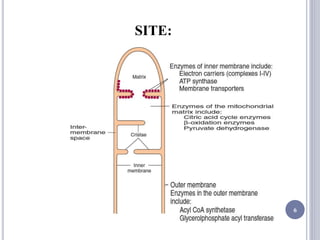
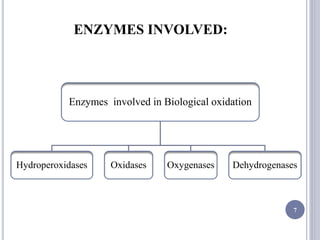
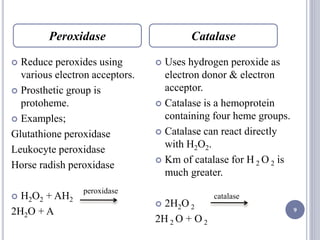
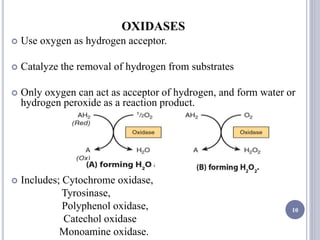
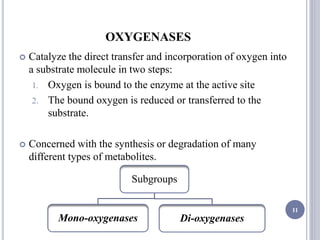
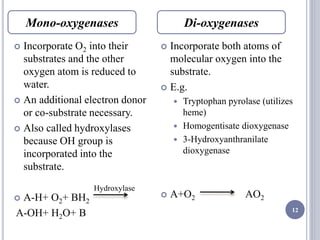
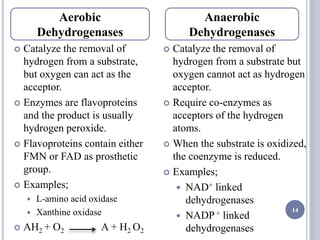
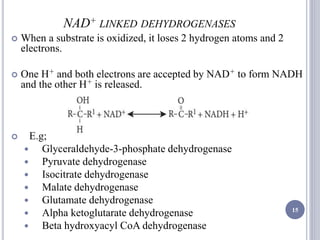
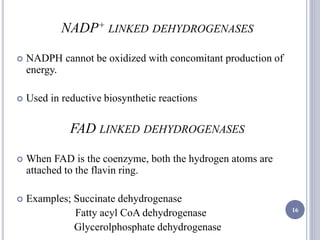
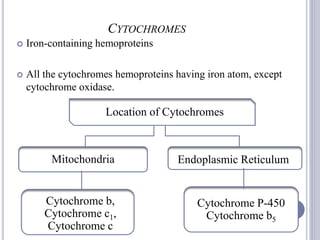
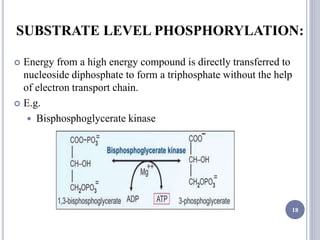
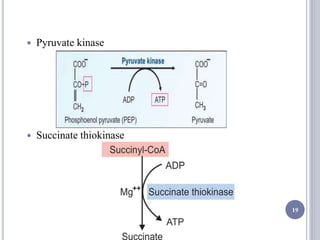
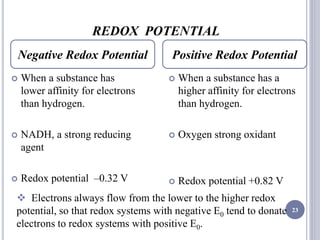
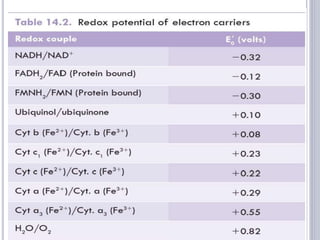
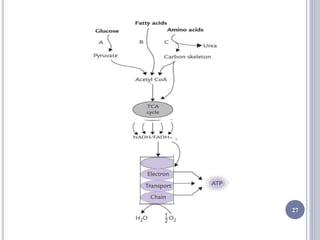
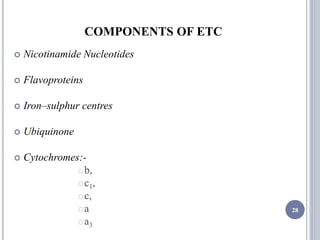
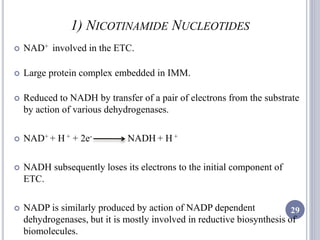
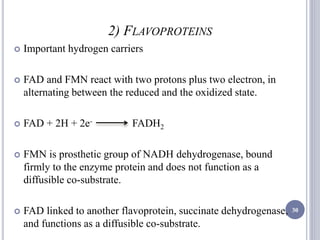
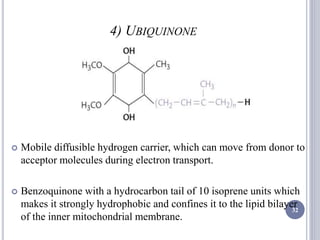
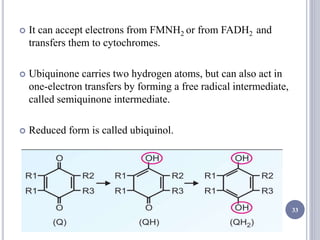
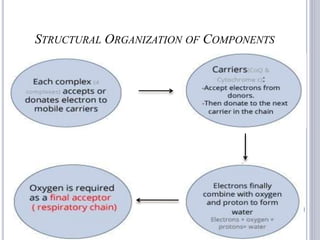
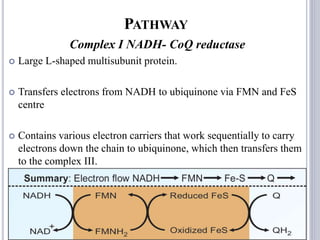
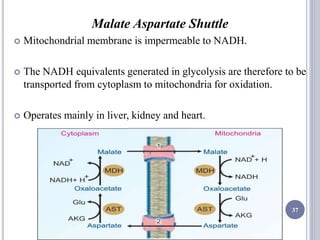
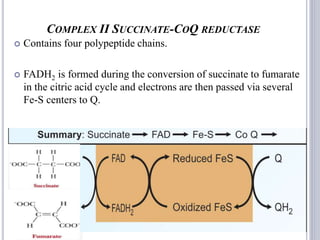
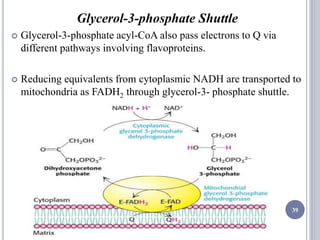
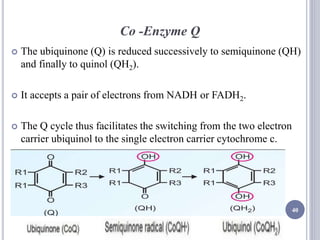
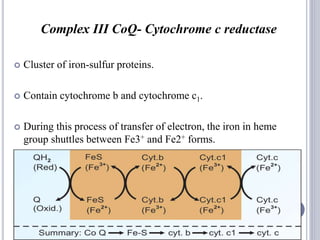
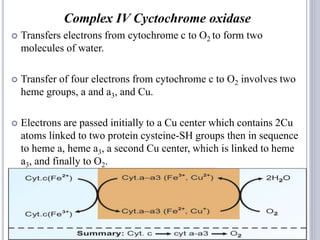
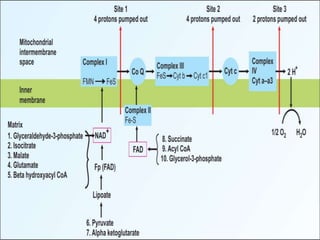
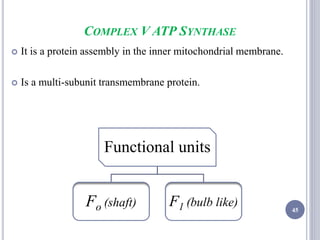
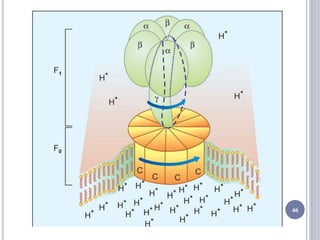
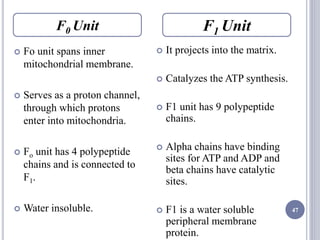
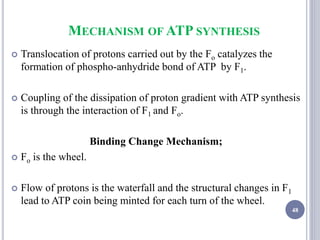
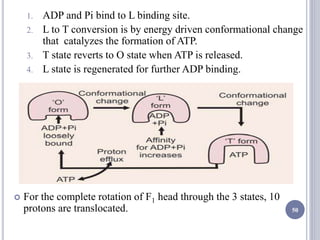
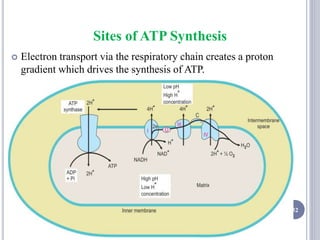
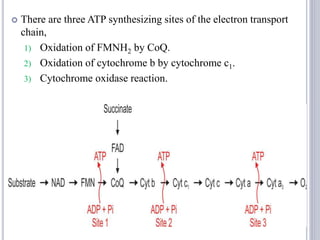
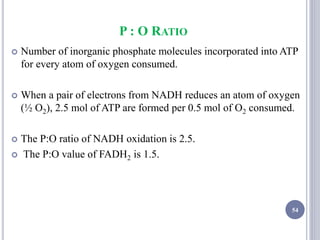
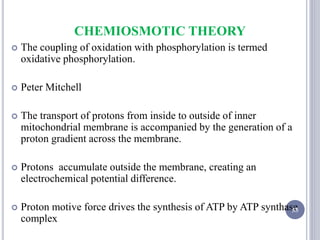
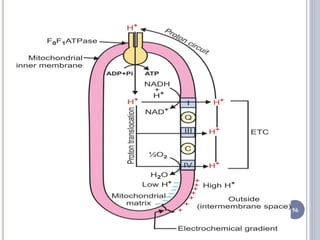
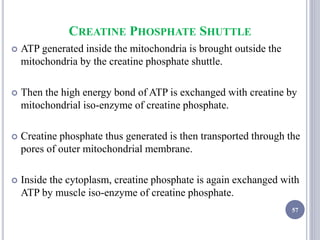
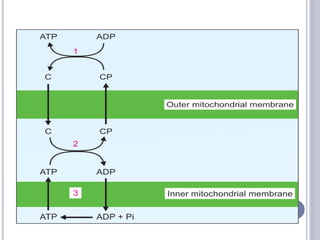
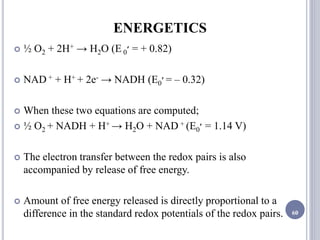
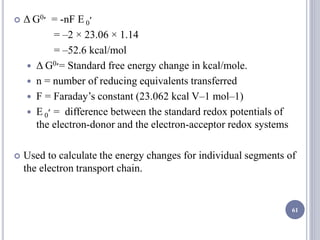
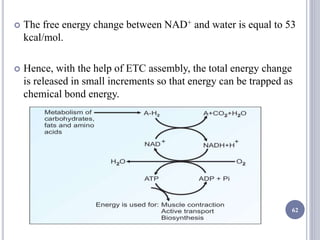
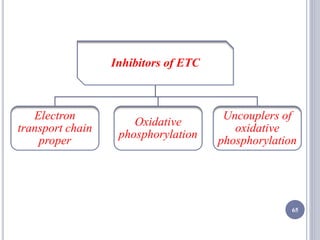
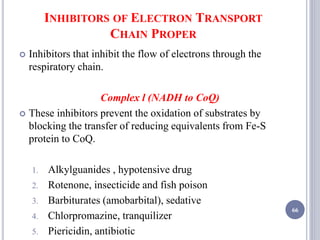
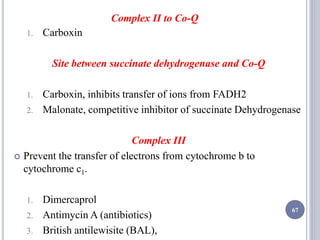
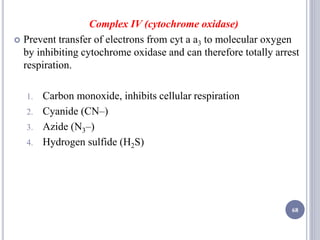
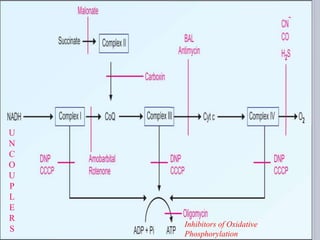
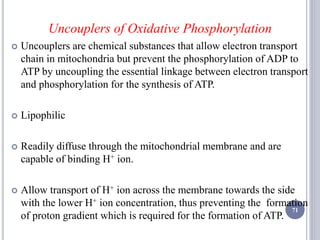
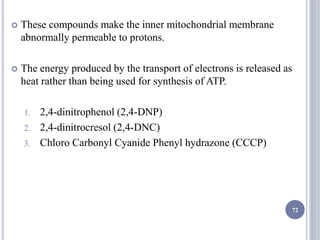
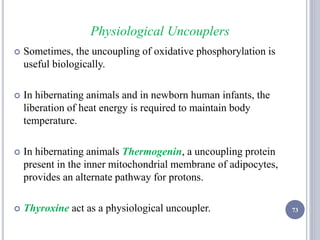
The document discusses biological oxidation and the electron transport chain, emphasizing the processes of oxidative phosphorylation and the roles of various enzymes and components involved in these reactions. It details stages of oxidation, enzyme classifications, and the mechanisms of ATP synthesis through the electron transport chain within mitochondria. Key concepts include redox potential, proton gradients, and the chemiosmotic theory, which describes how energy from electron transfer is harnessed to produce ATP.